Introduction – Understanding Proximity Extension Assay (PEA) Technology
Proximity Extension Assay (PEA) technology is a modern tool that allows scientists to measure many proteins at the same time, even in very small samples. This method is widely used in research laboratories, especially in studies that require highly sensitive and accurate protein detection.
PEA works by using pairs of antibodies that each attach to a specific part of a protein. Each antibody has a short piece of DNA linked to it. When both antibodies find and bind to the same protein, their DNA tags come close enough to interact. This interaction creates a new DNA sequence that can be copied and measured.
Because of this design, PEA has two major strengths:
High specificity – It only gives a signal when both antibodies bind the same protein.
High sensitivity – It can detect proteins present at very low levels in the sample.
Over the past decade, PEA has grown popular in biomarker discovery, proteomics research, and drug development studies (research use only). Many researchers choose it over older methods because it needs only a few microliters of sample and can measure hundreds or even thousands of proteins in one test.
In this article, we will explain:
- How PEA works step-by-step
- How it compares with other protein analysis tools like mass spectrometry
- Why it works so well with PCR amplification
- The difference between PEA and proximity ligation assay (PLA)
- The main advantages and research applications of this method
By the end, you will understand not only the principle of PEA technology, but also why it has become a go-to method for protein research in academic, biotech, and pharmaceutical laboratories.
The Principle of Proximity Extension Assay (PEA)
The proximity extension assay principle is built on the idea that two events must happen at the same time for a signal to be produced:
- Both antibodies must bind to the same target protein.
- The DNA tags on those antibodies must be close enough to interact.
This "two-key" requirement makes PEA very specific. It greatly reduces false signals caused by single antibodies sticking to the wrong protein.
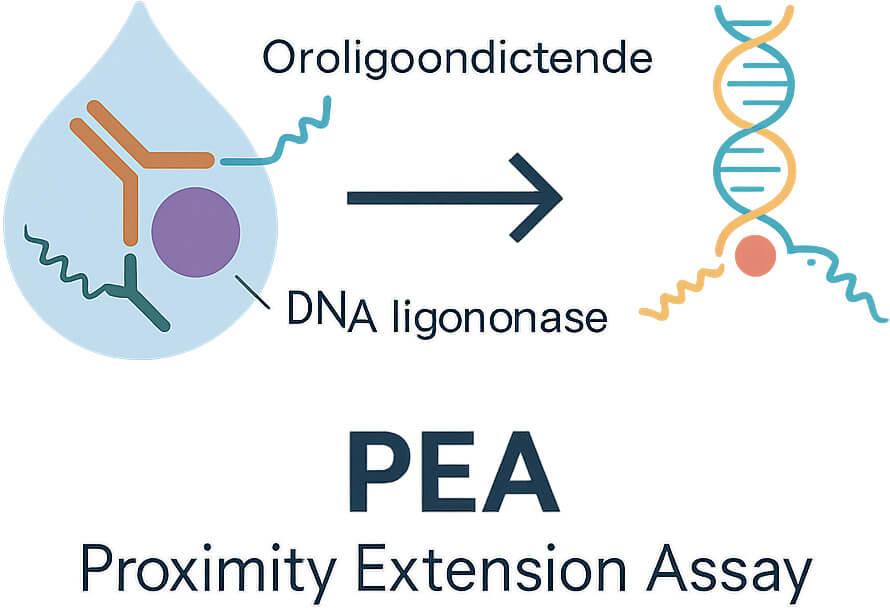
Step-by-Step Process
- Dual Antibody Binding – Each target protein is recognised by two different antibodies. One binds to one part (epitope) of the protein, and the other binds to another part.
- DNA Tag Proximity – Each antibody carries a short DNA strand. When both antibodies attach to the same protein, the DNA strands come close together.
- Hybridisation and Extension – The DNA strands are designed so that they can partially stick to each other (hybridise). A special enzyme, DNA polymerase, then extends one DNA strand using the other as a template, creating a brand-new DNA sequence.
- Signal Amplification – This unique DNA sequence is then copied many times using quantitative PCR (qPCR) or next-generation sequencing (NGS).
- Quantification – The strength of the DNA signal tells you how much of the target protein was in the sample. Data is expressed as NPX (Normalized Protein eXpression) values.
Why This Principle Works So Well
- Specificity: Both antibodies must bind, so random or non-specific binding is filtered out.
- Sensitivity: DNA-based amplification can detect even a handful of protein molecules.
- Multiplexing: Each protein has its own unique DNA barcode, allowing hundreds or thousands of proteins to be measured in one run.
In short, PEA turns protein detection into a DNA-reading problem — and since DNA can be copied and measured with extreme precision, the results are both sensitive and reliable.
PEA vs. Mass Spectrometry: Key Differences
Both PEA technology and mass spectrometry (MS) are powerful tools in proteomics research. However, they have different strengths and weaknesses depending on the research goals. Let's take a closer look at how these two technologies compare in important areas.
1. Sensitivity
PEA: Offers high sensitivity even for proteins present in very low amounts, detecting proteins with high specificity due to the dual-antibody binding mechanism. It works well with limited sample volumes (as low as 1–3 µL).
Mass Spectrometry: Also highly sensitive, but may struggle with extremely low-abundance proteins, especially in complex samples. MS works best when large amounts of sample are available (typically ≥50 µL).
2. Multiplexing Capability
PEA: One of the biggest strengths of PEA is multiplexing — the ability to measure hundreds or even thousands of proteins in a single experiment. This is possible because each protein gets a unique DNA barcode, allowing high-throughput assays in a single run.
Mass Spectrometry: While MS can measure hundreds of proteins, it often requires more complex and time-consuming sample preparation and analysis. In high-throughput studies, MS may require multiple runs to cover the full spectrum of proteins, limiting efficiency.
3. Sample Volume and Throughput
PEA: Requires a small sample volume (1–3 µL), which is ideal when working with limited or precious samples. Additionally, PEA assays are scalable, making them suitable for both small and large cohort studies.
Mass Spectrometry: Requires larger sample volumes (often ≥50 µL) and the process can be time-consuming, which can be a disadvantage in studies with limited sample availability or high-throughput needs.
4. Data Output and Quantification
PEA: Provides quantitative data in the form of NPX (Normalized Protein eXpression) values. These values are consistent and allow for easy comparison across different experiments and studies. The data is precise and reproducible, especially when paired with robust bioinformatics tools.
Mass Spectrometry: Also provides quantitative data, but the interpretation can be more complex. MS measures the abundance of peptides, which are then mapped to proteins. This process can introduce errors and variability, especially in samples with a complex protein profile.
5. Complexity and Ease of Use
PEA: Is generally easier to use than MS. The process involves simple antibody binding, DNA hybridisation, and PCR amplification. The results are easy to interpret, and the method is relatively straightforward to implement in any proteomics lab.
Mass Spectrometry: While MS is a powerful tool, it is more technically demanding and requires expert knowledge for both sample preparation and data interpretation. The equipment and protocols are more complex, and the system can be expensive to maintain.
6. Application Focus
PEA: Is particularly useful for targeted proteomics, where you are interested in quantifying a specific set of proteins. It excels in areas like biomarker discovery, drug development, and disease research where a known list of target proteins is studied.
Mass Spectrometry: Is better suited for untargeted proteomics — the discovery of unknown proteins. It's commonly used when researchers are trying to find new biomarkers or proteins that haven't been identified yet.
Comparison Table
| Feature | PEA Technology | Mass Spectrometry |
| Sensitivity | High sensitivity, even for low-abundance proteins | High sensitivity but may miss very low-abundance proteins |
| Multiplexing | Can analyze hundreds to thousands of proteins in one run | Typically analyzes fewer proteins, may need multiple runs |
| Sample Volume | Only 1–3 µL required | Requires ≥50 µL of sample |
| Data Output | Quantitative, reproducible NPX values | Quantitative data, but can be complex to interpret |
| Complexity | Easy to use and scalable | Requires specialized training and complex setup |
| Application Focus | Targeted proteomics, biomarker discovery | Untargeted proteomics, biomarker discovery |
PEA technology is particularly useful when researchers need highly reproducible, quantitative results from a predefined set of proteins, making it an ideal choice for targeted biomarker studies and clinical research (non-disease claims).
Proximity Extension Assay (PEA) and PCR – A Powerful Combination
One of the main reasons proximity extension assay technology is so sensitive is because it turns a protein detection problem into a DNA detection problem. DNA is much easier to copy and measure with precision, and this is where PCR (polymerase chain reaction) comes in.
Why PCR Works So Well with PEA
DNA barcodes for each protein – In PEA, every target protein is linked to a unique DNA sequence created when the two antibody tags come together and are extended by DNA polymerase.
Exponential amplification – PCR can make millions of copies of that DNA sequence from just a few starting molecules. This means even proteins present at extremely low levels can be detected.
High specificity – The PCR primers are designed to match the unique DNA barcode for each protein, so only the correct sequence is amplified.
Types of Readouts in PEA
qPCR (Quantitative PCR)
- Measures DNA amplification in real time.
- Ideal for medium-sized protein panels such as Olink Target 96, which profiles 92 proteins at once.
- Fast and cost-efficient for studies that focus on a fixed set of targets.
NGS (Next-Generation Sequencing)
- Reads all DNA barcodes in parallel, making it possible to profile thousands of proteins in a single run.
- Used in high-throughput formats such as Olink Explore 3072, which measures over 3,000 proteins.
- Enables large-scale studies with deep data coverage.
Because PCR amplifies only the DNA sequences created by the PEA process, background noise is kept low and results are reproducible. This combination of dual-antibody specificity and PCR amplification power is what makes PEA stand out in modern proteomics.
Applications of Proximity Extension Assay (PEA)
The proximity extension analysis (PEA) is used in many areas of scientific research. Here are some main applications:
1. Biomarker Discovery and Validation
PEA helps scientists find specific proteins—called biomarkers—that change in amount under different conditions. For example, researchers measured over 1,100 proteins in plasma and found six new proteins that differed in patients with severe brain injury (traumatic brain injury) compared to healthy controls.
2. Researching Molecular Pathways
Scientists study how proteins work together in pathways inside cells. Because PEA can measure many proteins at once from tiny samples, it is ideal for this type of research.
3. Drug Development and Monitoring
PEA allows researchers to see how drugs affect protein levels. It helps track a drug's impact by measuring changes in proteins before and after treatment, giving insights into how drugs work and their effects.
4. Studies with Limited Sample Material
PEA needs only a tiny amount of sample—often just a few microliters. This makes it a great choice for rare or precious samples such as cerebrospinal fluid, tumor biopsies, or dried blood spots.
5. High-Throughput Protein Profiling
Some versions of PEA can measure hundreds or even thousands of proteins from many samples at once. One study used a high-throughput workflow with PEA and sequencing to measure nearly 1,500 proteins in 96 samples, producing nearly 150,000 data points in a single run.
These applications make PEA a flexible and powerful tool in biomarker research, drug studies, and biological pathway analysis, especially when sample size is small and the research goal is clear.
Proximity Ligation Assay (PLA) vs. Proximity Extension Assay (PEA)
Both Proximity Ligation Assay (PLA) and Proximity Extension Assay (PEA) are advanced methods used to detect and measure proteins. While they share some similarities, they have key differences in how they work and what they're best used for.
How They Work
PLA: Involves two antibodies that bind to different parts of the same protein or to two proteins close together. These antibodies are linked to short DNA strands. When the antibodies are close enough, the DNA strands can be joined together by an enzyme called ligase. This creates a new DNA sequence that can be amplified and measured.
PEA: Also uses two antibodies, each attached to a unique DNA tag. When both antibodies bind to the same protein, the DNA tags come close enough to interact. This interaction allows a DNA polymerase enzyme to extend one DNA strand using the other as a template, forming a new DNA sequence that can be amplified and measured.
Key Differences
| Feature | PLA | PEA |
| Signal Generation | DNA ligation | DNA extension |
| Enzyme Used | DNA ligase | DNA polymerase |
| Sample Complexity | Can be affected by complex samples | More robust in complex samples |
| Multiplexing | Typically lower (up to ~96 proteins) | Higher (up to 3072 proteins) |
| Readout Methods | Fluorescence, microscopy, flow cytometry | qPCR, NGS |
When to Use Each
Use PLA when you need to study protein interactions within cells or tissues, as it allows for spatial localization and can be used in situ (directly in the sample).
Use PEA when you need to analyze many proteins simultaneously in complex biological samples, such as blood or plasma, especially when working with small sample volumes.
Advantages of Proximity Extension Assay (PEA)
The proximity extension assay technology (PEA) offers several key advantages compared to older laboratory methods. These benefits make it a powerful and popular choice for research scientists studying proteins.
1. Exceptional Sensitivity and Specificity
- Highly sensitive: PEA can detect extremely low levels of proteins, in the femtomolar range, thanks to the unique DNA-based amplification step.
- Highly specific: It only produces a signal when two antibodies bind the same protein, reducing false-positive results.
2. Superb Multiplexing from Tiny Sample Volumes
- PEA can measure hundreds to thousands of proteins at once, all from just 1 µL of sample.
- This is especially helpful when working with precious or scarce samples like cerebrospinal fluid or tumor biopsies.
3. Wide Dynamic Range
- It can detect proteins across a wide range of concentrations—up to 9 orders of magnitude—so both common and rare proteins are measured accurately.
4. Robust Performance in Complex Samples
- PEA works well even with complicated sample types like plasma or serum. It avoids interference that can occur with other assays, making the results reliable.
- High reproducibility in diverse samples strengthens confidence in the findings.
5. Simplified, High-Throughput Workflow
- The PEA process is performed in a homogeneous, single-tube format, which reduces errors and makes the method simpler than techniques that need many steps.
- It's ideal for high-throughput studies, where many samples are tested quickly and efficiently.
Summary Table
| Key Advantage | What It Means for Research Use |
| High sensitivity & specificity | Detects tiny amounts of protein accurately |
| Multiplexing from low volumes | Tests many proteins using very little sample |
| Wide dynamic range | Detects both common and rare proteins reliably |
| Robust for complex samples | Works well with messy biological fluids |
| Easy workflow, high throughput | Allows quick testing of many samples with fewer steps |
These strengths make PEA an excellent tool for biomarker discovery, drug development research, and proteomics studies (all for research use only—no clinical diagnosis or treatments). Its efficiency, accuracy, and scalability make it a favorite among scientists in academia, biotech, and pharmaceutical R&D.
Conclusion and Future Directions of Proximity Extension Assay (PEA)
Proximity Extension Assay (PEA) has revolutionized protein analysis by offering a highly sensitive, specific, and scalable method for detecting multiple proteins simultaneously from minimal sample volumes. Its unique combination of dual-antibody recognition and DNA amplification has made it a preferred choice for researchers in various fields, including biomarker discovery, drug development, and disease research.
Looking Ahead: Future Directions
As proteomics research continues to evolve, several advancements are on the horizon for PEA technology:
Integration with Next-Generation Sequencing (NGS): Combining PEA with NGS platforms allows for ultra-high multiplexing, enabling the analysis of thousands of proteins in a single experiment. This integration enhances the depth and breadth of proteomic studies, facilitating comprehensive biomarker discovery and validation.
Expansion of Protein Panels: Ongoing efforts aim to expand the number of proteins that can be detected using PEA, covering a broader range of the human proteome. This expansion will provide a more complete picture of protein expression and function in health and disease.
Advancements in Automation: The development of automated workflows for PEA assays will streamline the process, reduce hands-on time, and increase throughput. Automation will also improve consistency and reproducibility across experiments.
Enhanced Data Analysis Tools: The creation of more sophisticated bioinformatics tools will aid in the interpretation of complex data generated by PEA assays. These tools will help researchers identify meaningful patterns and correlations, accelerating the discovery of novel biomarkers and therapeutic targets.
Clinical Translation: While PEA is currently used primarily in research settings, its potential for clinical applications is being explored. With further validation and standardization, PEA could become a valuable tool for non-clinical research applications, aiding in disease diagnosis, prognosis, and monitoring.
Final Thoughts
PEA technology stands at the forefront of proteomic analysis, offering unparalleled sensitivity, specificity, and scalability. As advancements continue, PEA is poised to play a pivotal role in the future of biomedical research, providing deeper insights into the molecular mechanisms underlying health and disease.
Reference
- Homogeneous antibody-based proximity extension assays provide sensitive and specific detection of low‑abundant proteins in human blood (Nucleic Acids Research, 2011) — Demonstrates PEA achieves femtomolar sensitivity and covers a wide dynamic range using just 1 µL of plasma.
- Homogenous 96‑Plex PEA Immunoassay Exhibiting High Sensitivity, Specificity, and Excellent Scalability (PLoS ONE, 2014) — Describes a scalable 96‑plex PEA format, including performance in complex samples like plasma and dried blood spots. PLOS+1
- Development and validation of a quantitative Proximity Extension Assay instrument with 21 proteins associated with cardiovascular risk (PLoS ONE, 2023) — Shows the creation of a custom 21‑protein PEA panel with high sensitivity and precision for cardiovascular research.
- Proximity Extension Assay in Combination with Next‑Generation Sequencing for High‑throughput Proteome‑wide Analysis (Molecular & Cellular Proteomics, 2021) — Illustrates how PEA with NGS readout enables measurement of ~1,500 proteins across 96 samples in one batch.
- Proteomics by qPCR Using the Proximity Extension Assay (PEA) (Methods in Molecular Biology, 2025) — Provides a detailed, up-to-date protocol covering all practical and QC aspects of conducting PEA assays.




