- Overview
- Panels List
- Applications
- Workflow
- Why Creative Proteomics
- Demo
- Sample Requirements
- Case Study
- FAQ
- Sample Submission Pack
Why Focus on the Horse Proteome with Olink Panels?
The Olink-Horse Proteome Assay is optimized for equine-specific proteomic profiling. This platform addresses the unique challenges of equine biomarker discovery by combining high specificity for horse proteins with the scalability required for veterinary research and clinical applications.
- Exceptional Specificity for Equine Biomarkers – The dual-antibody recognition system ensures high specificity by requiring two independent antibodies to bind to the same target protein before DNA amplification. This minimizes false positives and enables accurate detection of low-abundance proteins in horse samples. While the core technology is designed for human proteins, Olink's platform can be adapted for equine studies through species-specific antibody validation, ensuring reliable measurement of horse-specific biomarkers critical for monitoring diseases.
- Ideal for Longitudinal and Population-Level Equine Studies – With the ability to profile hundreds of proteins simultaneously from minute sample volumes, the assay is uniquely suited for large-scale equine cohorts. This enables biobank-scale studies tracking proteomic changes across breeds, ages, or environments. It also allows Longitudinal monitoring of performance horses, capturing dynamic responses to training, stress, or therapeutic interventions.
- Enhanced Target Specificity Compared to Alternatives – While platforms like SomaScan rely on aptamer-based detection, Olink's antibody-based PEA technology demonstrates superior specificity for defined protein targets. This reduces off-target interactions and strengthens correlations between protein measurements and phenotypic traits in horses, which is critical for validating biomarkers in equine health and disease.
Selecting the Right Panel for Horse Proteome Studies
Mapping the equine proteome requires both breadth for discovery and precision for validation. We offer a tiered panel strategy designed specifically for horse-focused research:
- High-Dimensional Exploratory Discovery
The highest tier is defined by its large-scale, unbiased exploratory capacity for novel biomarker discovery, utilizing the highest-plex Explore 384 platform. This includes the Explore 384 Inflammation and Inflammation II panels, which comprehensively analyze 384 proteins each to provide a systems-level view of inflammatory pathways and networks, enabling the discovery of new biological insights without a pre-defined focus.
- Metabolic and Immunometabolic Interaction
This tier investigates the cross-talk between metabolic processes and immune function, a field known as immunometabolism. The Target 96 Metabolism panel resides here, profiling proteins that act as metabolic regulators and energy sensors, which can influence and be influenced by immune responses, thereby linking metabolic health to immune competence.
- Specialized Target 96 Immune & Inflammatory Pathways
This tier expands into deeper, more specialized immune and inflammatory mechanisms using the Target 96 platform, which offers a broader analysis of 96 proteins. It encompasses three distinct panels: the Target 96 Immuno-Oncology panel for profiling the tumor microenvironment and cancer immunity; the Target 96 Immuno-Response panel for broader adaptive and innate immune system activity; and the Target 96 Inflammation panel, which focuses on proteins involved in acute and chronic inflammatory processes. These panels are designed for hypothesis-driven research into specific immune and inflammatory pathways.
- Core Targeted Cytokine Profiling
This foundational tier is dedicated to focused analysis of a central set of signaling proteins, primarily cytokines and chemokines, which are crucial proteins for cell signaling and immune modulation, secreted by various cells to coordinate immune responses and inflammation. The Target 48 Cytokine panel serves this tier, providing a concentrated profile of 48 key cytokines to monitor essential immune communication and inflammatory status.
Together, this hierarchy progresses offer a structured framework within the context of horse biology for selecting the appropriate panel, ensuring high specificity, reduced cross-reactivity risks, and enhanced data quality.
Here is a list of recommended panels we offer:
Table. List of Olink Panels
| Platforms | Recommended Olink Panels |
| Explore 384 | Olink Explore 384 Inflammation |
| Olink Explore 384 Inflammation Ⅱ | |
| Target 96 | Olink Target 96 Inflammatory |
| Olink Target 96 Metabolism | |
| Olink Target 96 Immune Response | |
| Olink Target 96 Immuno-Oncology | |
| Target 48 | Olink Target 48 Cytokine |
Additional Options
In addition to the panels above, we can offer panels for human-focused research:
- Explore HT
- Explore 3072 (3072 proteins)
- Explore 1536 (1536 proteins)
- Explore 384
- Target 96
- Olink Target 96 Cell Regulation Panel
- Olink Target 96 Oncology II Panel
- Olink Target 96 Oncology III Panel
- Olink Target 96 Neuro Exploratory
- Olink Target 96 Organ Damage Panel
- Olink Target 96 Development Panel
- Olink Target 96 Cardiometabolic
- Olink Target 96 Cardiovascular II Panel
- Olink Target 96 Cardiovascular III Panel
- Olink Target 96 Neurology
- Custom Olink Flex systems
we also offer panels for mouse-focused research:
- Target 96
- Target 48
Our team can assist in designing an integrated Olink–Horse Proteome Assay strategy tailored to your project goals.
Applications of Horse Proteome Assay
Our Horse Proteome Assay Service is designed to support advanced research in equine health, performance, and disease using cutting-edge proteomic technologies. By leveraging species-specific assays and high-resolution mass spectrometry, this service enables deep profiling of the equine plasma, serum, and tissue proteomes for diverse applications:
- Doping Control and Athletic Performance Monitoring
Identify and validate plasma protein biomarkers associated with illicit substance administration in racehorses. Using label-free quantitative proteomics and data-independent acquisition (DIA) mass spectrometry, this service can detect exposure to prohibited agents. With high specificity for equine blood proteins, it supports ethical sports monitoring and minimizes false positives.
- Exercise Physiology and Metabolic Adaptation
Investigate protein expression changes in response to acute and chronic exercise in Thoroughbreds. Quantify proteins involved in energy metabolism, antioxidant activity, and oxygen transport to understand adaptations to endurance racing and high-intensity training. This application is critical for optimizing training regimens and improving performance.
- Inflammatory and Acute Phase Response Profiling
Monitor acute phase proteins (APPs) and inflammatory markers in serum, wound fluid, and tissues using absolute quantification via concatenated peptide standards (QconCAT) and selected reaction monitoring (SRM). This enables precise tracking of conditions like systemic inflammation, infections, and post-surgical recovery in horses, supporting veterinary diagnostics and therapeutic interventions.
- Gastrointestinal and Systemic Disease Biomarker Discovery
Identify salivary and plasma protein signatures associated with equine gastric ulcer syndrome (EGUS) and other common pathologies. Proteins such as adenosine deaminase (ADA), thioredoxin (TRX), and immunoglobulin heavy constant mu show differential expression in diseased states and respond to treatment, offering non-invasive diagnostic and monitoring tools.
- Multi-Omics Integration for Translational Equine Research
Combine proteomic data with genomic and transcriptomic profiles to uncover protein quantitative trait loci (pQTLs) and signaling pathways conserved across species. This systems biology approach enhances the translation of equine research to human athletic medicine, metabolic disorders, and inflammatory diseases while supporting drug target identification and biomarker validation.
Workflow of Horse Proteome Assay
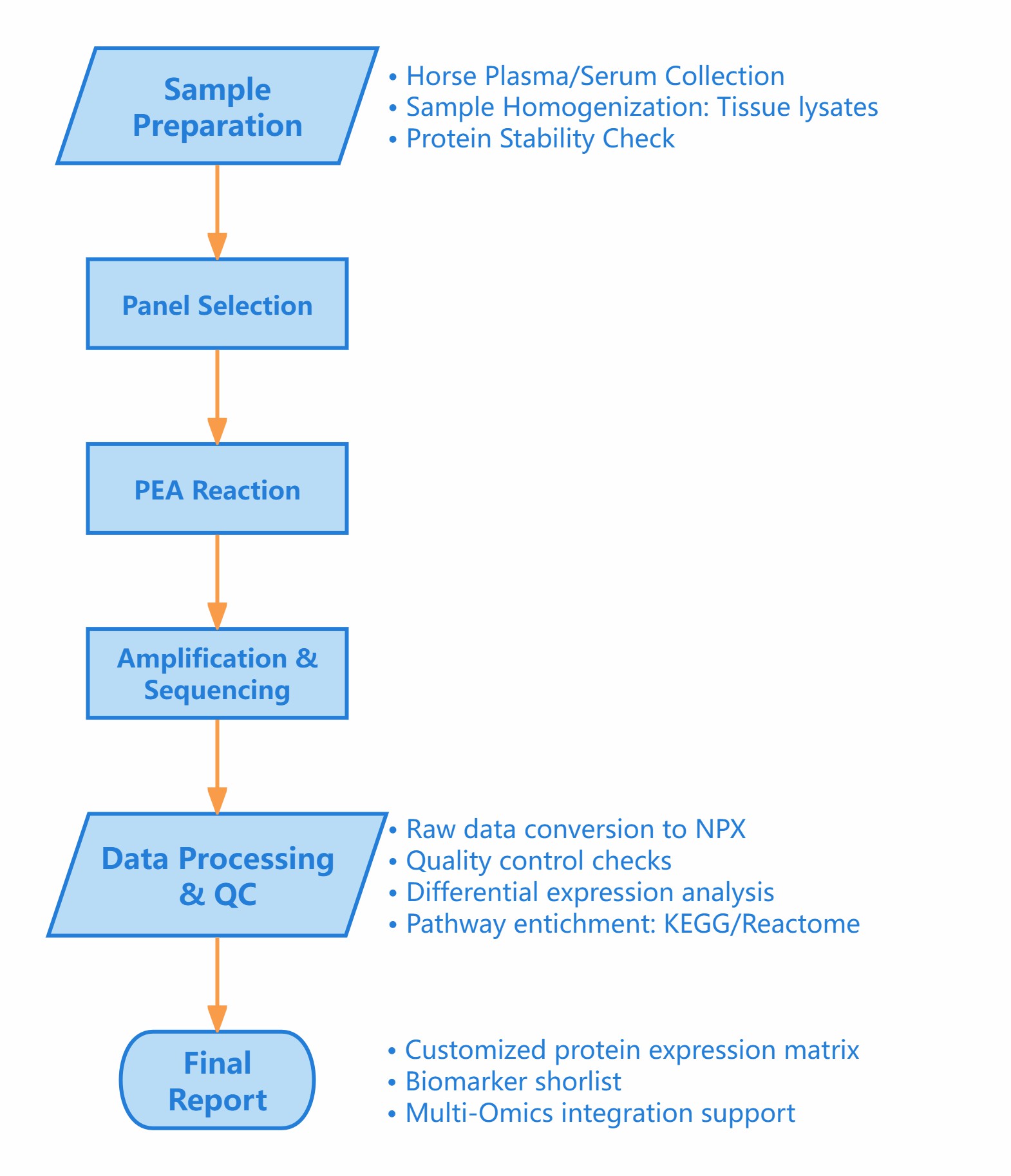 Figure 1: Workflow of Olink's Horse Proteome Assay Service.
Figure 1: Workflow of Olink's Horse Proteome Assay Service.
Why Creativ Proteomics
Olink's Horse-Specific Proteomics empower researchers to uncover robust, biologically relevant Insights in equine models. CPR offers a tailored solution for addressing unique challenges in equine health, performance, and translational research:
Horse-Optimized Assay Technology with Superior Performance
- Species-Specific PEA Technology: Our Olink's proprietary Proximity PEA platform is designed for high specificity and sensitivity in equine samples. This dual antibody-DNA binding mechanism minimizes cross-reactivity and enables accurate quantification of low-abundance proteins in complex matrices like horse serum, plasma, synovial fluid, and bronchoalveolar lavage (BAL) fluid.
- Comprehensive Equine Panels: Tailored panels provide focused coverage of equine-specific immune pathways, inflammation markers, musculoskeletal health indicators, and performance metabolism, enabling deep phenotyping in areas like exercise physiology, orthopedic disorders, and respiratory diseases.
Multi-Omics Integration for Mechanistic Insights in Equine Studies
- Proteogenomic Applications: Integrates seamlessly with genomic and transcriptomic data to identify protein quantitative trait loci (pQTLs) in horses, elucidating pathways relevant to athletic performance, metabolic disorders, and infectious diseases.
- Cross-Species Translational Relevance: Bridges horse-to-human research by highlighting evolutionarily conserved biomarkers and physiological pathways, enhancing the predictive value of equine data for human clinical applications in areas like musculoskeletal research and respiratory health.
Rigorous Quality Assurance for Equine Research
- Validation in Equine Samples: Assays are validated for precision, reproducibility, and dynamic range in equine-specific matrices, ensuring reliability in longitudinal studies, monitoring training adaptations, and multi-omics collaborations.
- Consistency in Low-Volume Samples: Maintains high data quality even with minute volumes (e.g., from precious clinical samples or limited serial collections from the same animal), reducing individual variability and supporting robust statistical analysis.
Workflow Efficiency and Customization for Equine Studies
- Custom Panel Flexibility: We offer tailored equine panels for hypothesis-driven validation or broad exploratory screening, aligning with diverse research needs from fundamental discovery to translational validation and veterinary diagnostic development.
- Integration with Equine Multi-Omics Platforms: Compatible with NGS and mass spectrometry workflows, enabling combined proteomic-genomic analyses without requiring highly specialized instrumentation, thus facilitating comprehensive studies on health, nutrition, and performance.
Together, Our Olink's horse-specific platform combines unmatched technical rigor, species-relevant design, and translational adaptability to address critical gaps in in equine and comparative preclinical research.
Demo Results: Olink Horse Proteome Assay
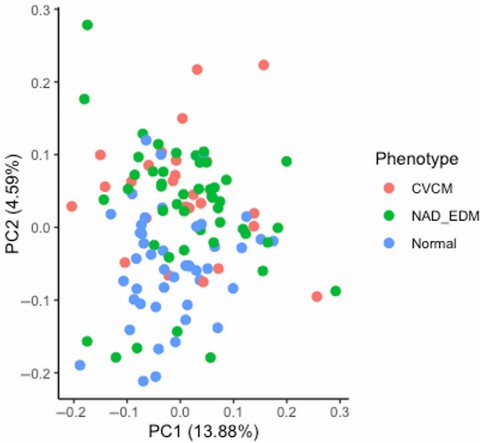 Figure 5: Principal component plot of CSF proteins showing the first two components (PC1 and PC2). (Donnelly C.G., et al. 2023)
Figure 5: Principal component plot of CSF proteins showing the first two components (PC1 and PC2). (Donnelly C.G., et al. 2023)
Sample Requirements for Horse Proteome Assay
1. Supported Sample Types
- Plasma/Serum: 15-20 μL (per panel).
- Urine: >100 μL (after conccentration).
- Tear Fluid: >50 μL.
2. Storage and Shipping
- Storage: -80°C immediately after collection.
- Shipping: Transport on dry ice (ensure no thawing during transit). Use validated shipping containers to maintain temperature.
Notes:
- Avoid repeated freeze-thaw cycles: This is critical to prevent protein degradation and loss of signal.
- Quick Processing: Process samples promptly after collection under cold conditions.
Case Study
Title: Cerebrospinal fluid and serum proteomic profiles accurately distinguish neuroaxonal dystrophy from cervical vertebral compressive myelopathy in horses
Journal: J Vet Intern Med
Year: 2023
- Background
- Methods
- Results
Spinal ataxia is a major neurological issue in horses, with Cervical Vertebral Compressive Myelopathy (CVCM) and equine neuroaxonal dystrophy/degenerative myeloencephalopathy (eNAD/EDM) being leading non-infectious causes. Antemortem differentiation between these conditions is extremely challenging. While an infectious cause like Equine Protozoal Myeloencephalitis (EPM) can be confirmed immunologically, no definitive diagnostic test exists for CVCM or eNAD/EDM. Diagnosis often relies on imaging techniques like radiography and myelography, which have poor sensitivity and cannot definitively diagnose eNAD/EDM. Consequently, a definitive diagnosis for many ataxic horses is only achieved post-mortem (necroscopy), creating a significant financial burden for owners. The study proposes that next-generation proteomic technologies, which have successfully identified biomarkers for human neurodegenerative diseases like Neurofilament-light (NEFL), could be applied to equine medicine to discover diagnostic protein profiles in serum and cerebrospinal fluid (CSF) that distinguish these conditions from each other and from normal horses.
The core proteomic technology employed was the Olink Proximity Extension Assay (PEA). The study utilized the Olink Explore 384 Neurology panel, a multiplex immunoassay designed to simultaneously measure the relative concentration of 368 neurologically relevant proteins using only 1μL of serum or CSF per sample.
The Olink PEA technology successfully characterized the proteomic profiles of equine serum and CSF. Quality control was passed for 86% (serum) and 89% (CSF) of individual assays, with an intra-assay coefficient of variation of 12%. Of the 368 proteins targeted, a subset was detectable in the equine samples: 146 proteins were detected in serum and 84 proteins were detected in CSF. Subsequent analysis revealed that 30 of the 146 serum proteins and 18 of the 84 CSF proteins were differentially abundant among the normal, CVCM, and eNAD/EDM groups after correction for multiple testing. While serum protein analysis did not show clear group separation, Principal Component Analysis (PCA) of the CSF data revealed a distinct clustering of samples from neurologically normal horses compared to those with spinal ataxia, though some overlap remained between the CVCM and eNAD/EDM groups.
Machine learning analysis of this high-dimensional Olink data identified a highly accurate diagnostic biomarker combination from the CSF. A conditional inference model selected a 2-protein test using CSF R-spondin 1 (RSPO1) and neurofilament-light (NEFL) as the most accurate classifier.
This model, as illustrated in Figure 3, discriminated the groups with the following accuracies: 87.2% for normal horses, 84.6% for CVCM, and 73.5% for eNAD/EDM. The violin plots in Figure 4 visually demonstrate the significant differences in abundance of these key proteins (RSPO1, NEFL) between the diagnostic groups.
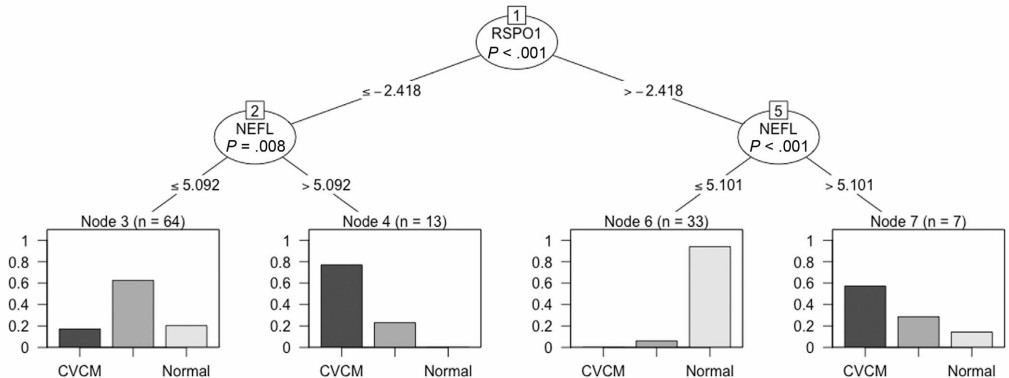 Figure : Conditional inference tree for CSF. (Donnelly C.G., et al. 2023)
Figure : Conditional inference tree for CSF. (Donnelly C.G., et al. 2023)
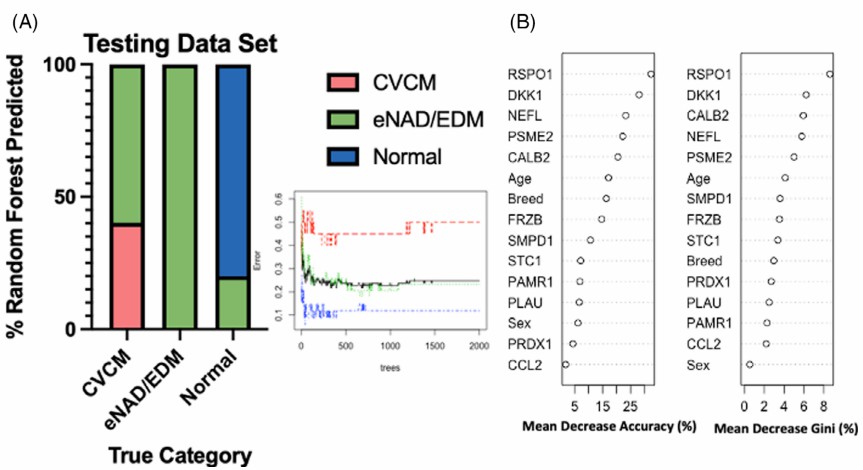 Figure 5: Random forest analysis of CSF. (Donnelly C.G., et al. 2023)
Figure 5: Random forest analysis of CSF. (Donnelly C.G., et al. 2023)
FAQs
How does Olink ensure quality control?
The platform incorporates rigorous QC steps, including:
- Internal controls within each assay.
- Incubation controls to monitor assay performance.
- Sample-specific data quality flags to identify potential outliers.
Are the results comparable to other platforms like ELISA?
While correlations can be strong for some proteins, results from PEA technology are not directly interchangeable with those from ELISA or other platforms (like aptamer-based SomaScan) due to fundamental differences in technology, antibody pairs, and detection methods. It is recommended to use the same platform for all samples within a study for consistent comparisons.
Reference
- Donnelly C.G., Johnson A.L., Reed S. et al. Cerebrospinal fluid and serum proteomic profiles accurately distinguish neuroaxonal dystrophy from cervical vertebral compressive myelopathy in horses. J Vet Intern Med. 37(2), 689-696(2023).