- Overview
- Panels List
- Applications
- Workflow
- Why Creative Proteomics
- Demo
- Sample Requirements
- FAQ
- Sample Submission Pack
Why Integrate Olink Proteomics with Metagenomics?
Olink employs Proximity Extension Assay (PEA) technology, converting protein abundance into quantifiable DNA signals. This achieves ultra-sensitive detection of proteins at fg/mL levels, outperforming mass spectrometry in low-abundance biomarker quantification. Metagenome sequencing utilizes shotgun sequencing of total microbial DNA, enabling species/strain-level identification and functional annotation of genes involved in metabolic pathways, antibiotic resistance, or virulence. The integration correlates Olink-measured host proteins with metagenome-derived microbial features to establish causal relationships. It reveals how microbial shifts directly influence host protein networks in diseases.
Olink Technology
Proximity Extension Assay (PEA)
1) Antibody pairs conjugate to DNA oligonucleotides.
2) Antibody pairs bind to target protein.
3) Upon co-localization, the DNA strands hybridize and extend into amplifiable templates.
4) Quantifiable DNA signals via qPCR or NGS.
Metagenome sequencing
1) Microbial sample collection.
2) DNA extraction.
3) Sequencing.
4) De novo assembly or align to reference genome.
5) Metagenome amplicon sequencing or metagenomics random shortgun sequencing.
Advantages
- Unparalleled sensitivity: Olink detects fg/mL proteins inaccessible to mass spectrometry.
- High-resolution microbial profiling: Strain-level differentiation.
- Multiplexing capacity: Enables population-scale studies with minimal sample volumes.
- Integrated data: Bridges host pathophysiology and microbial drivers.
Olink & Metagenome Platform: Selecting the Right Panel
Recommended Panels for Integrated Analysis
Panels we can offer are all technically compatible since they quantify human/mouse proteins, but priority should align with your biological focus. To scientifically select the appropriate panel for joint analysis, three core dimensions must be considered.
- First is the research objective. For instance, when studying the immune mechanisms of IBD, inflammation-related panels should be prioritized.
- Second are the characteristics of panel types. Explore panels are suitable for exploratory research but require larger sample sizes, while Target series are better for targeted validation.
- Third are practical constraints, such as sample volume and budget.
For most host-microbiome studies, immune-inflammatory panels and metabolic panels are most directly relevant.
Table. List of Olink Panels
| Research Foucus | Recommended Olink Panels |
| Hosy-microbe Immunity | Explore 384 Inflammation Explore 384 / Inflammation Ⅱ |
| Target 96 Immune Response | |
| Target 96 Inflammatory | |
| Target 48 Cytokine | |
| Metabolic Disorder | Explore 384 Cardiometabolic Explore 384 Cardiometabolic Ⅱ |
| Target 96 Metabolism | |
| Target 96 Cardiometabolic | |
| Gut-Brain Axis | Explore 384 Neurology Explore 384 Neurology Ⅱ |
| Target 96 Neuro Exploratory | |
| Target 96 Neurology | |
| Therapeutic Response | Target 96 Immuno-Oncology |
| Immuno-Oncology Panel | |
| Unbiased Discovery | Explore 3072 (3072 proteins) |
| Explore 1536 (1536 proteins) |
Additional Options
In addition to the panels above, we also offer:
- Explore HT
- Explore 3072 / Explore 384
- Target 96 / Target 48
- Olink Target 96 Development Panel
- Olink Target 96 Organ Damage Panel
- Olink Target 96 Cell Regulation Panel
- Olink Target 96 Oncology II Panel
- Olink Target 96 Oncology III Panel
- Olink Target 96 Cardiovascular II Panel
- Olink Target 96 Cardiovascular III Panel
- Olink Target 96 Mouse Exploratory Panel
- Olink Target 48 Mouse Cytokine Panel
- Custom Olink Flex systems
Our team can assist in designing an integrated Olink–metagenome strategy tailored to your project goals.
Applications
This service excels in mechanistic disease research and therapeutic development:
Host-Microbe Interactions in Disease Mechanisms
- Cardiovascular & Metabolic Diseases: Gut microbiota-derived metabolites regulate host proteins linked to inflammation, thrombosis, and metabolism.
- Inflammatory Bowel Disease (IBD): Microbial dysbiosis modulates systemic immune responses via cytokine/chemokine signaling.
Biomarker Discovery for Medical Research
- Predictive Biomarkers for Therapy Response: Plasma immune checkpoint proteins measured by Olink Immuno-Oncology panels correlate with gut microbiome composition and response to ICB therapy. In psoriasis, Olink Inflammation panels identified STAMBP and chemokines as early indicators of paradoxical eczema post-biologic therapy.
- Diagnostic/Prognostic Biomarkers: Olink Cardiometabolic/Inflammation panels detected 15 dysregulated proteins linked to disease severity.
Diet-Microbiome-Host Protein Crosstalk
- Dietary interventions reshape the microbiome, altering host protein expression.
Therapeutic Development & Monitoring
- Antibiotic Impact Assessment: Antibiotic-induced dysbiosis disrupts bile acid metabolism, leading to dysregulation of liver metabolic proteins and epigenetic regulators.
- Probiotic/Prebiotic Efficacy: Olink & metagenomics quantifies changes in immune/metabolic proteins after microbiome-targeted interventions.
Multi-Omics Integration in Complex Diseases
- Neurological Disorders: Gut microbes produce neuroactive metabolites that modulate neuronal proteins via the gut-brain axis.
- Chronic Fatigue Syndrome (ME/CFS): Reduced microbial butyrate synthesis, leading to decreased plasma butyrate, end with elevated neuroinflammatory proteins.
Workflow of Olink & Metagenome
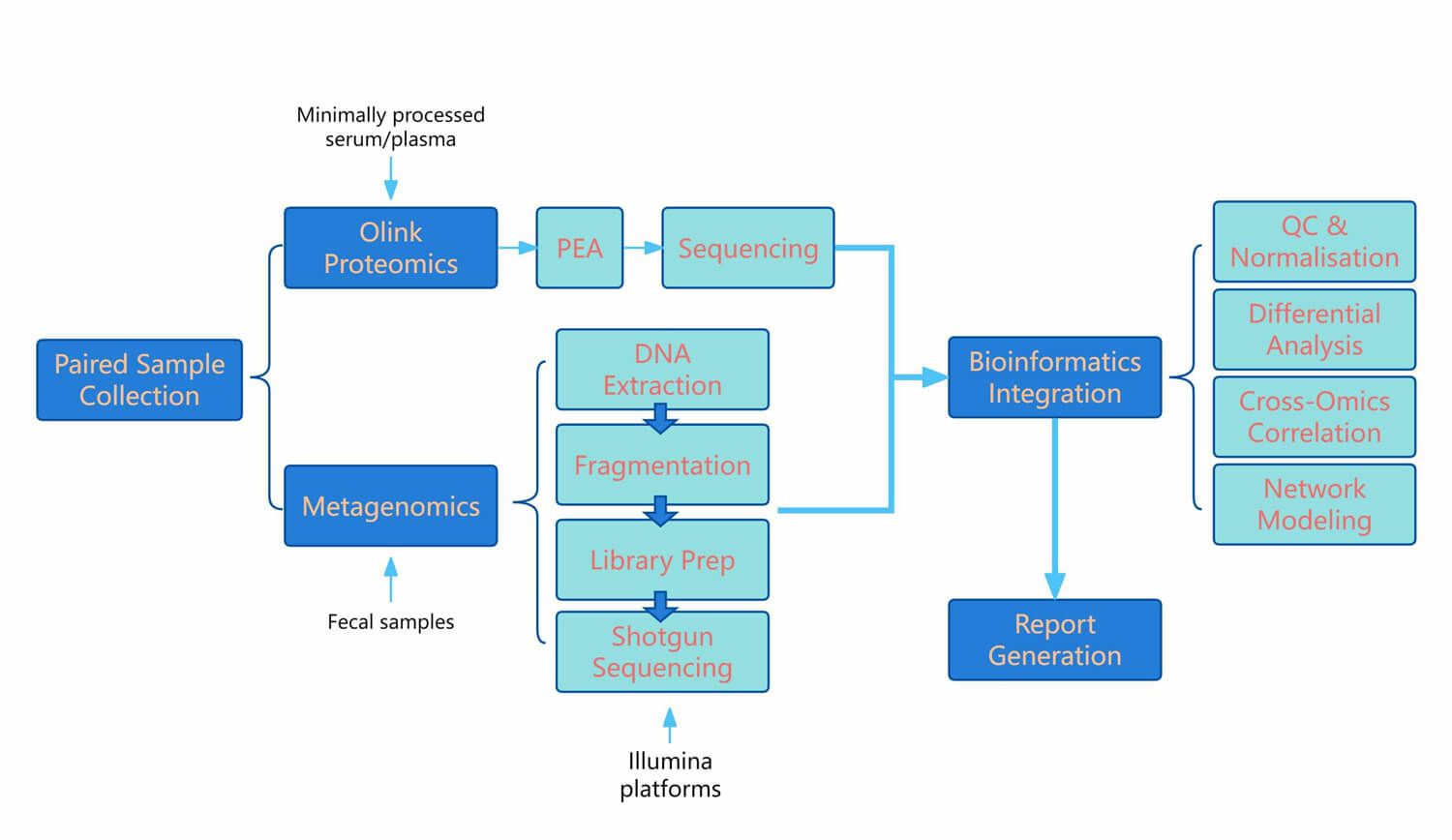 Figure 1: Workflow of integrated Olink proteomics and metagenome sequencing.
Figure 1: Workflow of integrated Olink proteomics and metagenome sequencing.
Why Creativ Proteomics
Unmatched Technical Performance and Sensitivity
- Ultra-low Sample Requirements: Enable high-precision protein quantification using minimal sample volumes. This is critical for rare or pediatric samples.
- High Sensitivity and Specificity: Detect low-abundance proteins with minimal cross-reactivity, outperforming traditional methods.
- Scalability: Olink Explore HT allows simultaneous profiling of >5,300 proteins from thousands of samples.
Integrated Multi-Omics Capabilities
- Synergistic Data Integration: Combine proteomic data (Olink) with metagenomic microbial composition, functional genes, and resistance markers.
- Cross-Platform Validation: Use orthogonal methods to enhance data reliability.
Expertise in Complex Study Design and Quality Control
- Rigorous Biobank-Scale Standards: Adherence to protocols from large cohorts like UK Biobank and China Kadoorie Biobank (CKB), ensuring reproducibility.
- Diverse Sample Proficiency: Validate for challenging matrices, with strict pre-analytical controls to minimize batch effects.
Advanced Bioinformatics
- Knowledge Graphs for Biomarker Discovery: Platforms contextualize protein-metagenome interactions, accelerating target identification.
Translational Impact and Customization
- Support drug target validation, microbiome-based therapies, and companion diagnostics.
- Tailored Solutions: Customized Olink panels.
Demo Results
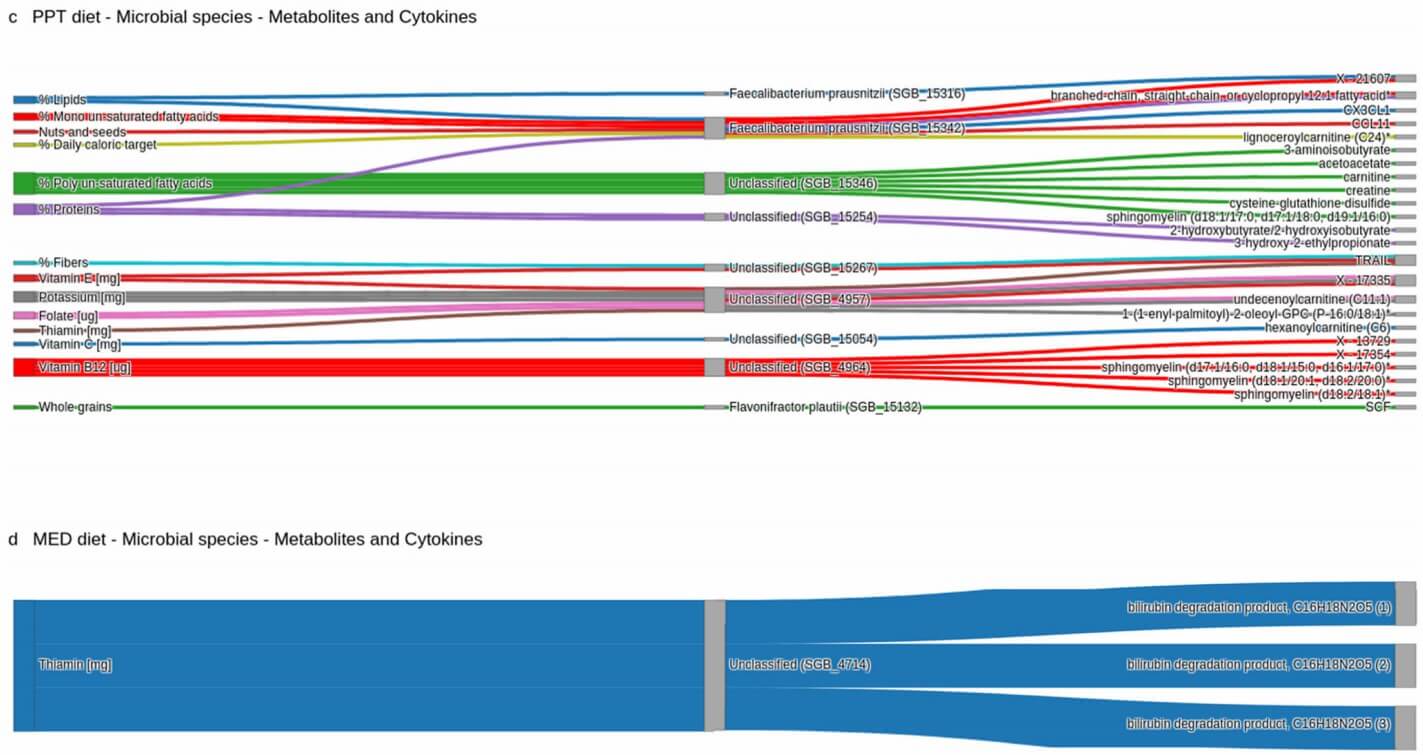 Figure 1: The mediatory effects of the microbiome were observed in all tested cytokines in the PPT group but in none of the tested cytokines in the MED group. (Shoer, S., et al. 2023)
Figure 1: The mediatory effects of the microbiome were observed in all tested cytokines in the PPT group but in none of the tested cytokines in the MED group. (Shoer, S., et al. 2023)
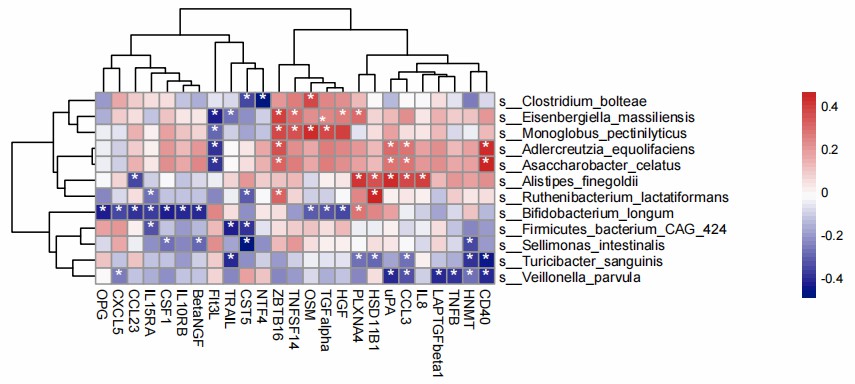 Figure 2: Heatmap of 25 serum proteins in IBD patients with at least one significant (q < 0.1) association with metagenomic taxonomic features. (Lee, JWJ., et al. 2021)
Figure 2: Heatmap of 25 serum proteins in IBD patients with at least one significant (q < 0.1) association with metagenomic taxonomic features. (Lee, JWJ., et al. 2021)
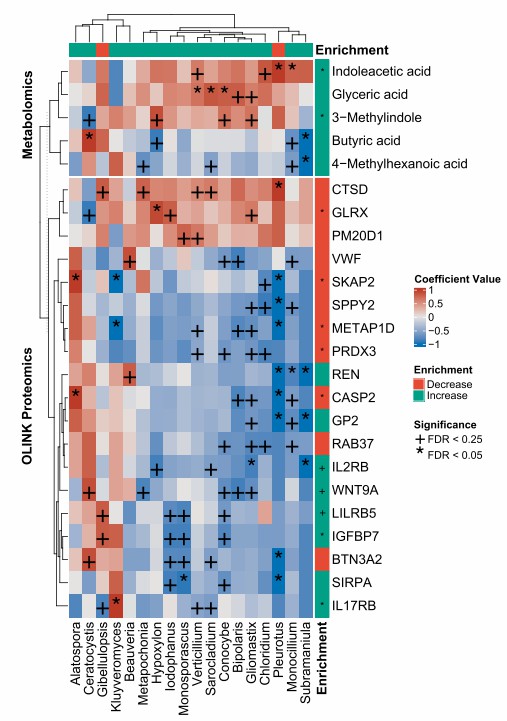 Figure 3: The correlations between changes in gut mycobiome and serum metabolome signatures, circulating proteomics profiles. (Wang, Y., et al. 2024)
Figure 3: The correlations between changes in gut mycobiome and serum metabolome signatures, circulating proteomics profiles. (Wang, Y., et al. 2024)
Sample Requirements
- Olink Proteomics sample type: Plasma, serum, body fluid, tissue, cells, exosomes.
- Sample size: 40 µL/sample at least.
- Protein concentration: 0.5mg/ml ~ 1mg/ml.
- Metagenome sequencing sample: ≥200mg of fecal material preserved in stabilizing buffers.
- Pre-treatment and Storage: Aliquot using sterile containers, preserve at -80°C.
- Always use paired samples from the same subject.
FAQs
How do Olink PEA technology and metagenomics integrate experimentally?
Olink uses proximity extension assays to convert protein detection into NGS-readable DNA sequences. It allows simultaneous processing with metagenomic libraries on platforms like Illumina NovaSeq.
Metagenome sequencing enables high-accuracy microbial genome assembly. This complements Olink' s protein quantification for host-microbe interaction studies.
Can both analyses run on the same sample?
Serum/plasma samples are ideal for Olink. For metagenomics, stool/tissue samples are required.
For plasma samples, it is theoretically possible to use the same sample for two types of analyses, but aliquoting is required. Divide the sample into portions, with one portion used for Olink assays and another for DNA extraction and metagenomic sequencing.
This approach is typically not feasible for stool samples or similar specimen types.
What are critical sample guidelines?
Olink analysis necessitates serum or plasma volumes as low as 1–2 µL. Samples should be stored at -80°C to prevent degradation, with strict avoidance of repeated freeze-thaw cycles to maintain protein integrity. And it supports diverse matrices including CSF and tissue lysates.
Metagenomics relies on stool or tissue samples preserved in DNA shield buffer at -80°C, with DNA extraction ideally completed within 24 hours of collection to minimize microbial composition shifts.
Both workflows mandate rigorous pre-analytical quality checks to ensure batch effect minimization and data reproducibility across large cohorts.
References
- Shoer, S., Shilo, S., Godneva, A., et al. (2023). Impact of dietary interventions on pre-diabetic oral and gut microbiome, metabolites and cytokines. Nature Communications, 14(1):5384. https://doi.org/10.1038/s41467-023-41042-x
- Lee JWJ., Plichta D., Hogstrom L., et al. (2021). Multi-omics reveal microbial determinants impacting responses to biologic therapies in inflammatory bowel disease. Cell Host Microbe, 29(8):1294-1304. https://doi.org/10.1016/j.chom.2021.06.019
- Wang Y., Chen J., Ni Y., et al. (2024) Exercise-changed gut mycobiome as a potential contributor to metabolic benefits in diabetes prevention: an integrative multi-omics study. Gut Microbes, 16(1):2416928. https://doi.org/10.1080/19490976.2024.2416928