- Overview
- Panels List
- Applications
- Workflow
- Why Creative Proteomics
- Demo
- Sample Requirements
- FAQ
- Sample Submission Pack
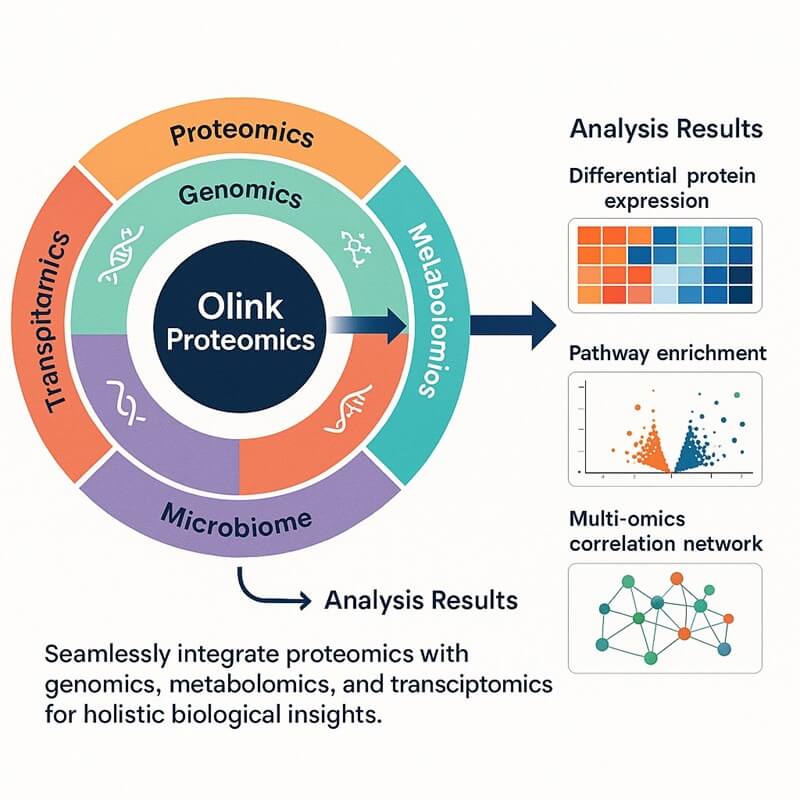
Why Integrate Olink Proteomics with scRNA-seq?
The integration of Olink and single-cell transcriptome (scRNA-seq) technologies operates on the principle of synergistic complementarity and cross-validation. scRNA-seq identifies cell subsets, transcriptional networks, and pathway-level alterations, while Olink simultaneously validates functional protein biomarkers within the same biological context. This dual-framework allows transcriptomic predictions to be rigorously verified at the protein level, thereby reducing false positives and establishing robust, biologically relevant correlations between gene expression and functional proteome dynamics.
Olink Technology
Proximity Extension Assay (PEA)
1) Antibody pairs conjugate to DNA oligonucleotides.
2) Antibody pairs bind to target protein.
3) Upon co-localization, the DNA strands hybridize and extend into amplifiable templates.
4) Quantifiable DNA signals via qPCR or NGS.
Single-Cell Transcriptome
1) Single-Cell Isolation and Capture: Cells are separated from tissues into suspensions.
2) Cell Lysis and RNA Barcoding: Captured cells are lysed to release mRNA.
3) Reverse Transcription (RT) and cDNA Synthesis: mRNA is reverse-transcribed into barcoded cDNA using the bead-bound primers.
4) cDNA Amplification and Library Preparation: cDNA is PCR-amplified to generate sufficient material for sequencing.
5) High-Throughput Sequencing: Libraries are sequenced, generating millions of short reads.
6) Data Processing.
Advantages
- Minimal Sample Requirements: Olink uses ≤6 µL of plasma; scRNA-seq requires from thousands to tens of thousands of cells.
- Unprecedented Sensitivity and Specificity: Olink detects low-abundance proteins missed by mass spectrometry, while scRNA-seq resolves rare cell populations.
- High-Precision Biomarker Discovery: Reduce false positives by cross-validating RNA-protein correlations.
- Comprehensive Pathway Mapping: Identify master regulators by linking transcriptional states to protein activity.
Olink & scRNA-seq Platform: Selecting the Right Panel
Recommended Panels for Integrated Analysis
The selection of an Olink panel for integration with scRNA-seq primarily depends on your research objectives and the specific biological insights you aim to capture. The reasons we recommend the following panels are:
- The Olink Explore HT panels, Explore 3072 panels, Explore 1536 panels are ideal for large-scale studies, analyzing thousands of proteins simultaneously to give a broad proteome context for scRNA-seq data.
- The Target 96 panels offer deep, focused coverage of specific biological pathways, suitable for targeted investigations cell subpopulations or pathways identified by scRNA-seq.
- The Target 48 Cytokine and Mouse Cytokine panels are crucial for studying immune dynamics, inflammation, and animal models, complementing scRNA-seq by detecting protein-level cytokine activity.
- Customized Olink Flex panels allow building custom assays based on scRNA-seq findings for precise protein analysis, enhancing integration of protein and transcriptome data.
Here is a list of recommended panels we offer:
Table. List of Olink Panels
Additional Options
In addition to the panels above, we also offer:
- Explore 384
Our team can assist in designing an integrated Olink–scRNA-seq strategy tailored to your project goals.
Applications
The integrated application of Olink proteomics and scRNA-seq enables researchers to uncover novel molecular interactions, dynamic regulatory networks, and system-level phenotypes across diverse biological contexts:
Deciphering Cellular Heterogeneity in Complex Tissues
- By combining scRNA-seq for unbiased cell-type classification with Olink' s high-plex protein quantification, researchers can resolve subtle cellular subpopulations and their functional proteomic signatures.
Mapping Cell Differentiation Trajectories and Lineage Commitment
- scRNA-seq-derived pseudotime trajectories can be validated against time-resolved protein activity using Olink. This reveals post-transcriptional regulatory nodes that drive fate decisions in stem cells or organoids.
Intercellular Signaling and Spatial Organization
- Integration enables reconstruction of ligand-receptor networks: scRNA-seq predicts cell-type-specific ligand/receptor gene pairs, while Olink quantifies secreted ligands in microenvironmental niches. Spatial validation is achieved via correlation with spatial transcriptomics or multiplexed imaging.
Dynamic Stress Responses and Cellular Plasticity
- Researchers track real-time adaptations to perturbations by coupling scRNA-seq transcriptional states with Olink-measured stress-response proteins. This identifies master regulators of plasticity, such as NRF2-mediated antioxidant responses.
Multi-Omics Network Inference and Functional Annotation
- Joint analysis identifies causal RNA-protein links. Pathway enrichment prioritizes high-impact nodes in disease-relevant networks.
Evolution of Protein Complexes and Allosteric Regulation
- Olink's PEA detects co-localized proteins in complexes, while scRNA-seq reveals co-expression of subunits. This uncovers context-dependent assembly, such as stress-induced remodeling of spliceosome complexes.
Workflow of Olink & scRNA-seq
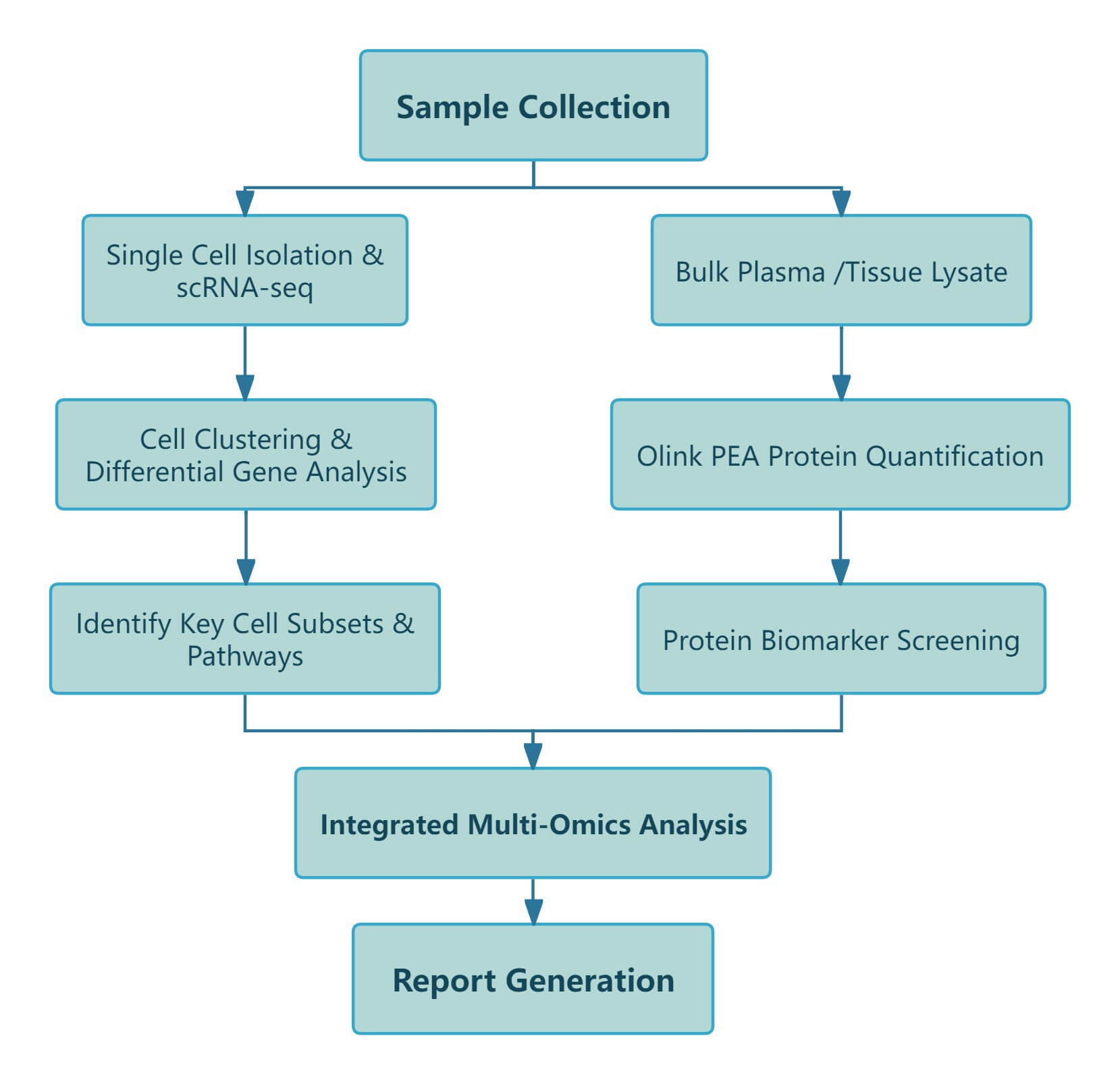 Figure 1: Workflow of integrated Olink proteomics and scRNA-seq.
Figure 1: Workflow of integrated Olink proteomics and scRNA-seq.
Why Creativ Proteomics
Cutting-Edge Technology Platform
- High-throughput capabilities: Support for large-scale projects (e.g., Olink Explore 3072 for >5,300 proteins and scRNA-seq platforms handling millions of cells).
- Rigorous validation: Compliance with certified standards, ensuring data reproducibility across batches and platforms.
End-to-End Process Optimization and Quality Control
- Minimal Sample Requirements: Ability to generate high-quality data from ultra-low inputs (e.g., Olink' s sensitivity with 1-6 µL of body fluids; scRNA-seq compatibility with frozen/rare samples.
- Standardized Workflows: Reduction of technical variability via robotics and embedded QC checkpoints.
- Cross-platform Compatibility: Seamless integration with major instrumentation.
Integrated Multi-Omics Analysis Expertise
- Unified Data Interpretation: Advanced bioinformatics tools to correlate protein (Olink) and gene expression (scRNA-seq) data, revealing biological interactions beyond single-layer insights.
- Customizable Analytics: Support for complex analyses.
Demo Results
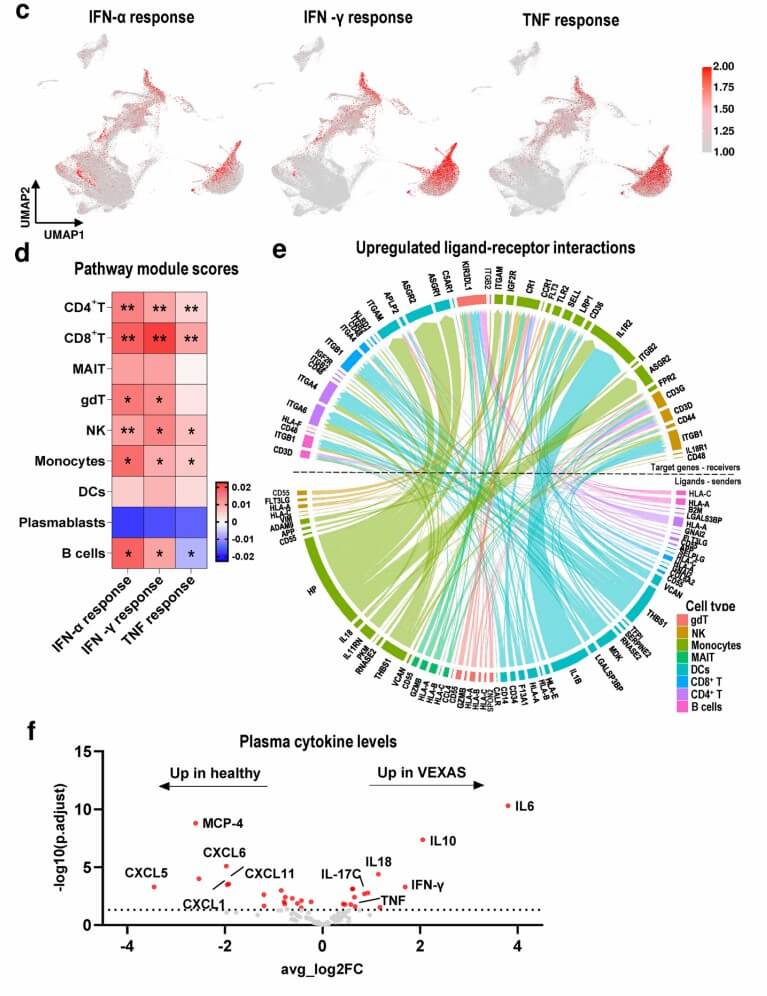 Figure 2: Compositional changes and activation of the inflammatory pathways in immune cells from VEXAS patients. (Mizumaki, H., et al. 2025)
Figure 2: Compositional changes and activation of the inflammatory pathways in immune cells from VEXAS patients. (Mizumaki, H., et al. 2025)
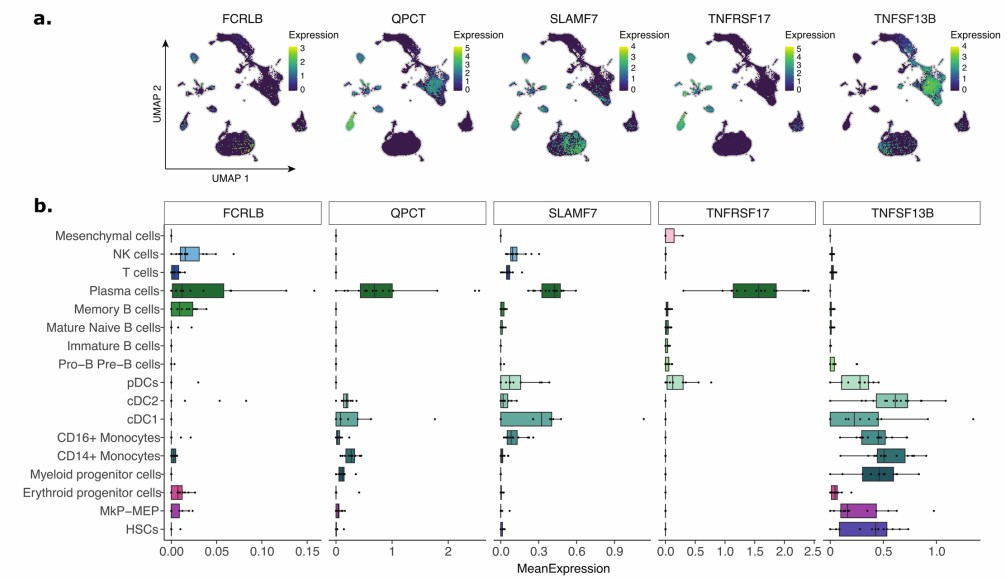 Figure 3: Gene expression levels of predictor proteins within the bone marrow (BM) immune ecosystem. (Carrasco-Zanini, J., et al. 2024)
Figure 3: Gene expression levels of predictor proteins within the bone marrow (BM) immune ecosystem. (Carrasco-Zanini, J., et al. 2024)
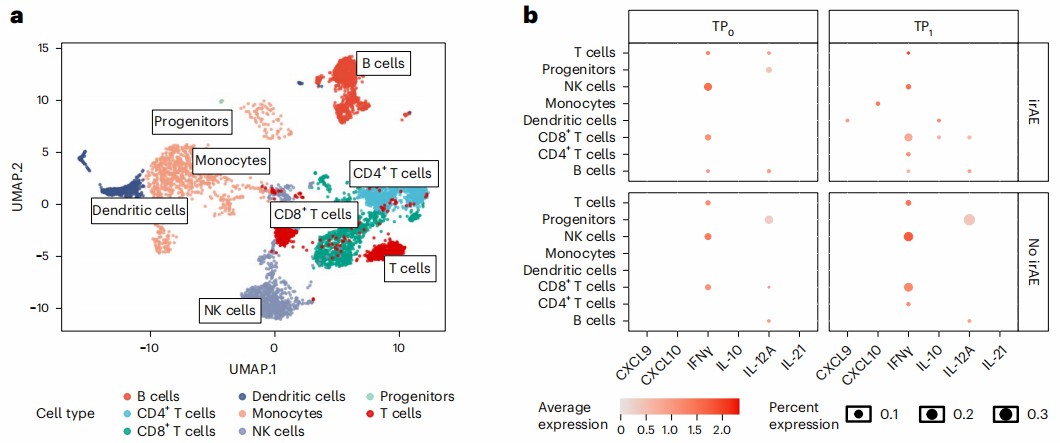 Figure 4: The dot plot illustrates the average expression of significantly altered cytokines (identified by MSD assay) in scRNA-seq-defined immune cell clusters at baseline (TP₀) versus irAE onset (TP₁) across six patients. (Dimitriou, F., et al.2024)
Figure 4: The dot plot illustrates the average expression of significantly altered cytokines (identified by MSD assay) in scRNA-seq-defined immune cell clusters at baseline (TP₀) versus irAE onset (TP₁) across six patients. (Dimitriou, F., et al.2024)
Sample Requirements
| Step | Olink Requirement | Metabolomics Requirement |
| Volume | Ultra-low input (1-6 µL for plasma/serum; 20-40 µL for CSF/urine) | ≥50 ng DNA post-extraction |
| Sample Types |
|
|
| Storage Temperature | -80°C (for long term), avoid freeze-thaw cycles | Cryopreserve in 90% FBS + 10% DMSO at -150°C (for long term); Preserve in cold buffer on wet ice(for short term) |
| Transport | Dry ice shipment; ≤-60°C | On dry ice or in validated cryopreservation media |
Notes
- Use split aliquots from the same donor/tissue. Fresh cells for transcriptomes; Plasma/lysate for proteomics.
FAQs
Can Olink detect intracellular proteins alongside scRNA-seq?
Yes. Olink technology is capable of detecting intracellular proteins and has been successfully integrated with scRNA-seq using methods such as SPARC. The SPARC method integrates modified Smart-seq2 scRNA-seq with Olink PEA to co-profile 89 intracellular proteins and global mRNA in single cells. The research extracts intracellular proteins through lysis buffer and verifies the concordance between protein data and mRNA data in cell differentiation trajectory analysis.
How to select Olink panels for scRNA-seq integration?
Olink panel selection should align with the biological context of the scRNA-seq study. Panels that directly match the investigated pathways or processes are preferred. For immune activation or inflammatory disease studies, choose the Inflammation or Immune Response panels; for metabolic or neurological research, select disease-specific panels like Cardiometabolic or Neurology. Custom-targeted panels are also available for hypothesis-driven studies requiring specific biomarkers.
How to integrate Olink protein data with scRNA-seq clusters?
First, scRNA-seq defines cell subtypes through clustering. Next, cluster-specific gene signatures are correlated with bulk protein levels from Olink assays in the same samples. This identifies functional protein biomarkers tied to specific cell states. For example, in obesity-linked osteoarthritis, scRNA-seq uncovered distinct synovial fibroblast subtypes, and Olink found elevated subtype-linked cytokines like FOS in activated fibroblasts. Key tools include CellChat, which maps ligand-receptor interactions between clusters, and the OlinkAnalyze R package. The latter uses ANOVA or linear mixed-effects regressions to validate protein-mRNA links.
Can Olink replace validation methods like ELISA or Luminex?
Yes, Olink can largely replace traditional methods such as ELISA or Luminex in the discovery phase of studies. This is owing to its high concordance, multiplex capability, and minimal sample volume requirements. However, for definitive validation, particularly when spatial context or cell-specific localization is critical, orthogonal methods like immunofluorescence, immunohistochemistry, or single-cell proteomics remain essential to complement Olink' s bulk-level protein quantification and ensure comprehensive biological verification.
References
- Mizumaki, H., Gao, S., Wu, Z. et al. In depth transcriptomic profiling defines a landscape of dysfunctional immune responses in patients with VEXAS syndrome. Nat Commun 16, 4690 (2025). https://doi.org/10.1038/s41467-025-59890-0
- Carrasco-Zanini, J., Pietzner, M., Davitte, J. et al. Proteomic signatures improve risk prediction for common and rare diseases. Nat Med 30, 2489–2498 (2024). https://doi.org/10.1038/s41591-024-03142-z
- Dimitriou, F., Cheng, P.F., Saltari, A. et al. A targetable type III immune response with increase of IL-17A expressing CD4+ T cells is associated with immunotherapy-induced toxicity in melanoma. Nat Cancer 5, 1390–1408 (2024). https://doi.org/10.1038/s43018-024-00810-4