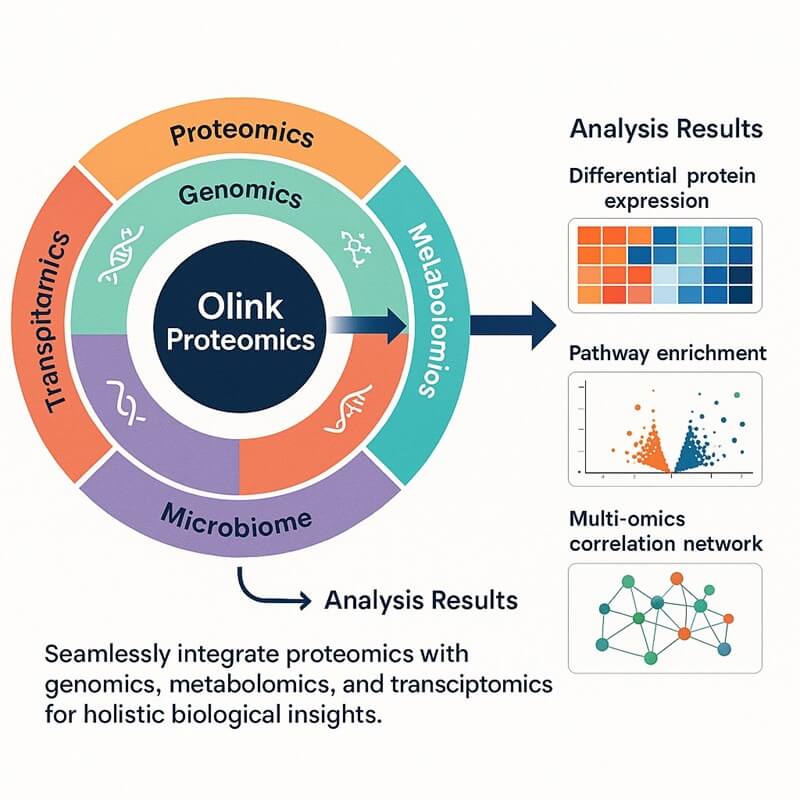
- Overview
- Panels List
- Applications
- Workflow
- Why Creative Proteomics
- Demo
- Sample Requirements
- FAQ
- Sample Submission Pack
Why Integrate Olink Proteomics with Metabolome?
The integration of Olink and metabolome allows researchers to map out complex layers of biological regulation together. Olink employs PEA technology to detect trace cytokines missed by conventional proteomics. While metabolomics captures real-time functional readouts. The integrated approach excels in translational efficiency. Olink' s microvolume compatibility preserves samples for parallel metabolomics, and optimized pipelines scale for large cohorts. Together, these techniques provide more cost-effective biological insights than either one alone could achieve. This integrated framework drives mechanistic discovery in biomedicine, transforming fragmented omics data into actionable networks of disease causality.
Olink Technology
Proximity Extension Assay (PEA)
1) Antibody pairs conjugate to DNA oligonucleotides.
2) Antibody pairs bind to target protein.
3) Upon co-localization, the DNA strands hybridize and extend into amplifiable templates.
4) Quantifiable DNA signals via qPCR
Metabolome
1) Sample Introduction: Solid/liquid samples are vaporized to enter the ion source as a gas.
2) Ionization: Gas-phase molecules are ionized.
3) Acceleration: Ions are accelerated by an electric field.
4) Deflection: Ions traverse a magnetic field, bending their paths.
5) Detection & Data Analysis: Ions are counted. m/z values are used to determine molecular weight, elemental composition, and fragmentation patterns.
Advantages
- Unparalleled Sensitivity: Olink detects fg/mL proteins inaccessible to mass spectrometry.
- Enhanced Biological Insight: Reveal causal protein-metabolite interactions, bridging upstream signaling with downstream metabolic dysregulation.
- Cost Efficiency: Microvolume compatibility allows multi-omics profiling from limited biopsies/biobank samples.
- Comprehensive Metabolite Coverage: Untargeted metabolomics detects metabolites via dual-column LC-MS/MS. While targeted panels enable precise quantification of specific pathways .
Olink & Metabolome Platform: Selecting the Right Panel
Recommended Panels for Integrated Analysis
Olink Panels are selected based on their direct relevance to metabolic pathways and disease mechanisms. Panels like Cardiometabolic, Metabolism, Diabetes, and Cytokine measure proteins directly involved in or regulated by metabolic processes, providing mechanistic links to metabolomic data.
- The Explore HT panels provide broad, hypothesis-free coverage of proteins linked to core metabolic processes.
- Target 96 panels (notably Metabolism, Development, Immuno Response, and Inflammatory) offer focused analysis of key pathways regulating nutrient sensing, energy metabolism, and immune-metabolic crosstalk.
- The Target 48 Cytokine panels capture cytokines critical for inflammation-driven metabolic dysregulation.
- Customized Flex panels enable disease-specific investigation of clinically actionable protein-metabolite interactions.
Here is a list of recommended panels we offer:
Table. List of Olink Panels
Additional Options
In addition to the panels above, we also offer:
- Explore 1536 (1536 proteins)
- Explore 384
- Target 96 / Target 48
- Olink Target 96 Cardiometabolic
- Olink Target 96 Neuro Exploratory
- Olink Target 96 Neurology
- Olink Target 96 Immuno-Oncology
- Olink Target 96 Organ Damage Panel
- Olink Target 96 Cell Regulation Panel
- Olink Target 96 Oncology II Panel
- Olink Target 96 Oncology III Panel
- Olink Target 96 Cardiovascular II Panel
- Olink Target 96 Cardiovascular III Panel
- Olink Target 96 Mouse Exploratory Panel
- Custom Olink Flex systems
Our team can assist in designing an integrated Olink–Metabolome strategy tailored to your project goals.
Applications
This service excels in mechanistic disease research and therapeutic development:
Biomarker Discovery and Validation
- Proteins and metabolites serve as direct readouts of cellular function and biological condition. By simultaneously analyzing changes in both proteins and metabolites, researchers can gain more comprehensive insights into the disease development process and underlying mechanisms.
Research on Disease Mechanisms
- Cardiovascular Disease: Olink-metabolome integration identifies causal protein-metabolite interactions driving vascular pathology.
- Cancer: Olink & Metabolome technology streamlines the search for therapeutic targets, advances proteomics research, and reveals mechanisms of tumorigenesis.
- Brain Disease: Proteomics and multi-omics, including brain imaging techniques, have facilitated the detection of therapeutic targets associated with brain disorders. Integrating omics and imaging data can yield novel insights for understanding brain diseases.
- Neuropsychiatric Disorders: Studies revealed dysregulated neuro-immune-metabolic crosstalk in conditions like PTSD.
Multi-Omics Mechanistic Discovery
- The approach uncovers regulatory axes across biological layers.
Patient Stratification Research
- The technology stratifies heterogeneous diseases into molecularly defined subtypes.
Workflow of Olink & Metabolome
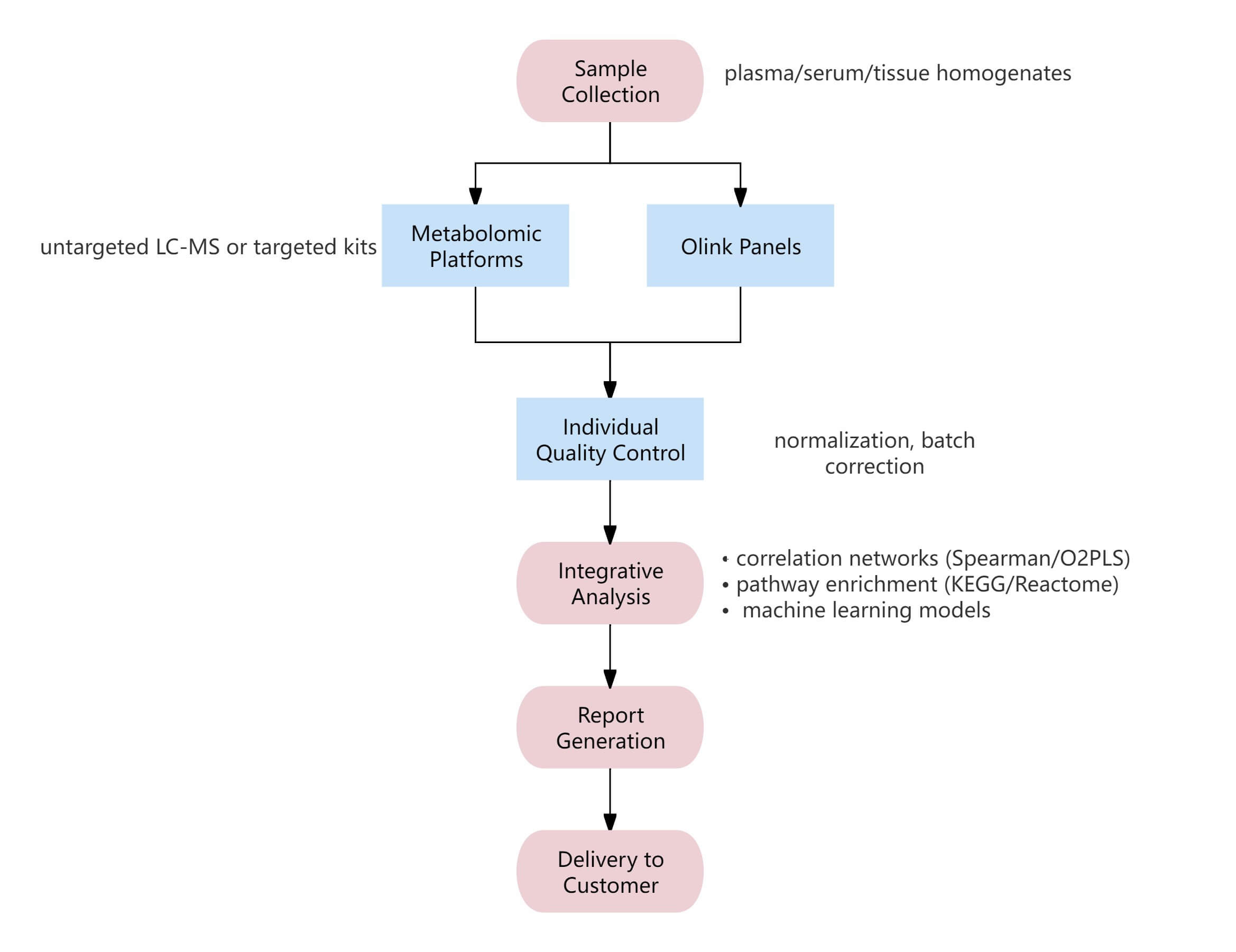 Figure 1: Workflow of integrated Olink proteomics and metabolome.
Figure 1: Workflow of integrated Olink proteomics and metabolome.
Why Creativ Proteomics
Multi-Omics Integration Capabilities
- Technical Complementarity: Olink' s PEA technology enables precise detection of low-abundance proteins (sensitivity down to fg/ml level). While metabolomics excels at capturing dynamic changes in small-molecule metabolites.
- Microvolume Compatibility: Olink requires only 1–6 µL of bodily fluid to detect thousands of proteins. Metabolomics must maintain comparable low-sample consumption to minimize sample depletion and enhance data consistency.
High-Precision Detection
- Olink' s PEA technology employs antibody-DNA conjugates for "dual recognition", eliminating cross-reactivity common in traditional multiplex assays.
- Metabolite Coverage requires reference libraries to ensure accurate identification and quantification.
Standardized Workflows
- End-to-end standardized pipelines from sample preparation to data analysis ensure cross-platform data comparability.
Multi-Omics Association Analytics
- Pathway-Level Integration: Maps Olink' s signaling pathway coverage to metabolic pathways (e.g., KEGG) to reconstruct "protein-metabolite" regulatory networks.
- Causal Inference Tools: Leverages pQTL and mQTL data to prioritize disease-driving targets.
Scalable Technology Platform
- Support flexible combinations of Olink products (Target 96/Explore 3072) with untargeted/targeted metabolomics to accommodate varying research scales.
Demo Results
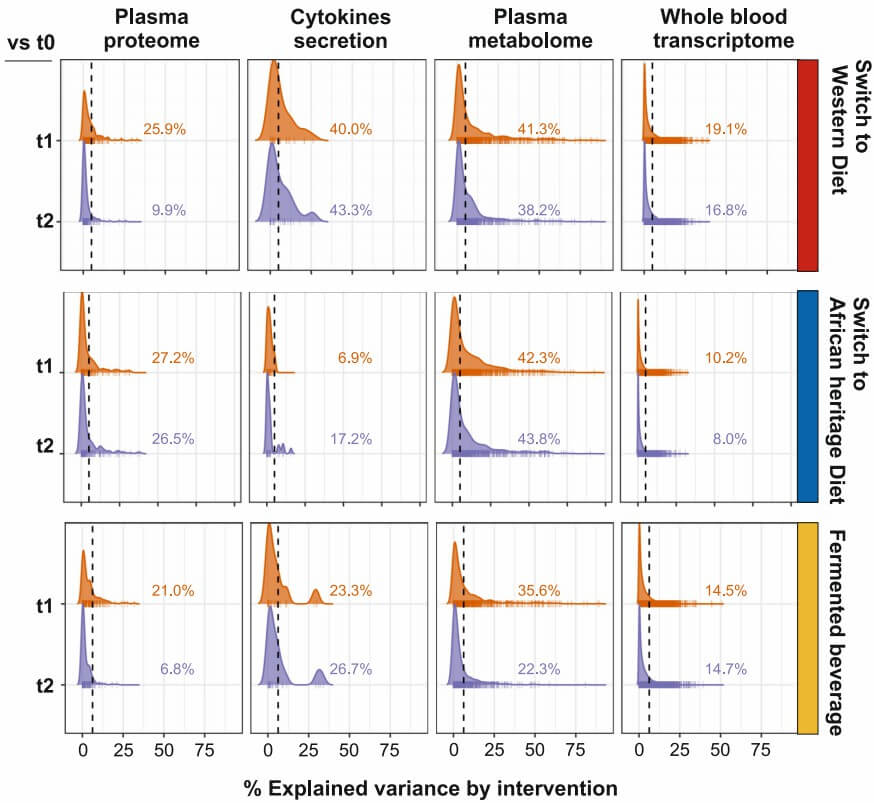 Figure 2: Percentage of explained variation attributable to diet intervention across multi-omics layers. (Temba, G.S., et al. 2025)
Figure 2: Percentage of explained variation attributable to diet intervention across multi-omics layers. (Temba, G.S., et al. 2025)
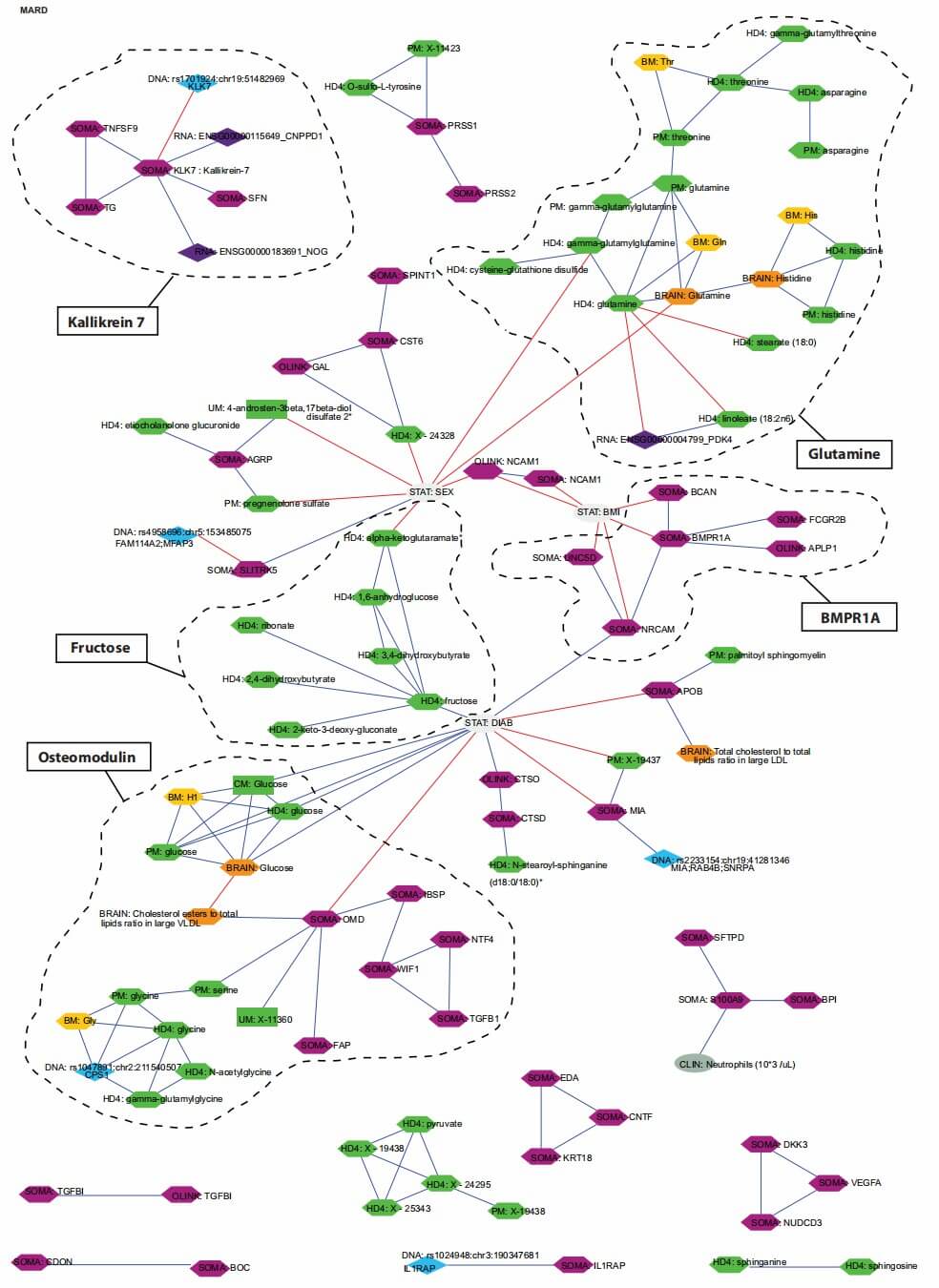 Figure 3: Multiomics interaction network of protein and metabolite signatures in MARD-associated clusters. (Halama, A., et al. 2024)
Figure 3: Multiomics interaction network of protein and metabolite signatures in MARD-associated clusters. (Halama, A., et al. 2024)
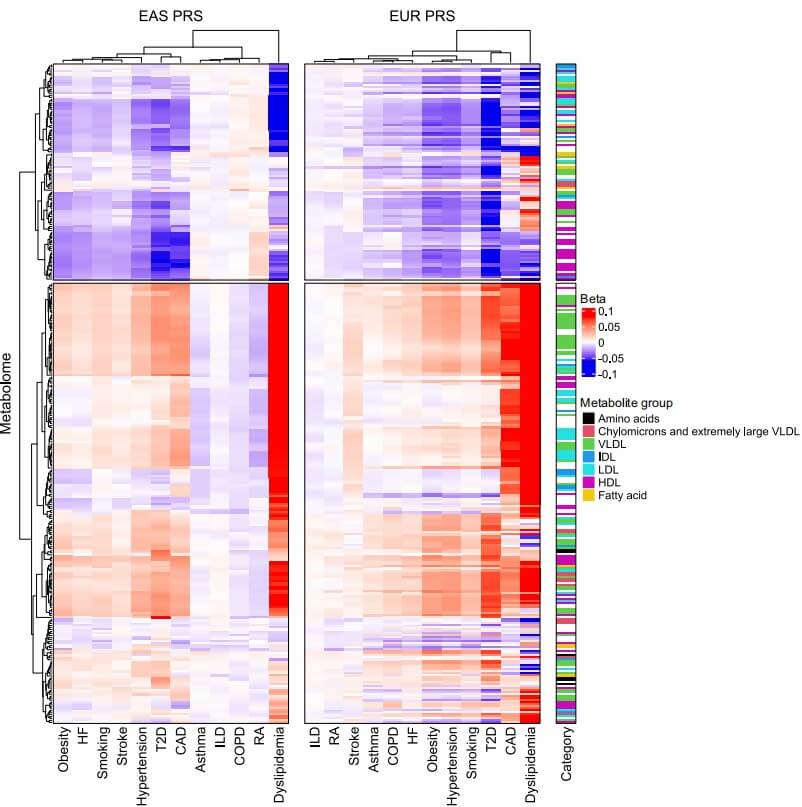 Figure 4: Genome-wide PRS associations with circulating NMR-metabolized lipid/metabolite markers. (Yamamoto, Y., et al. 2025)
Figure 4: Genome-wide PRS associations with circulating NMR-metabolized lipid/metabolite markers. (Yamamoto, Y., et al. 2025)
Sample Requirements
| Step | Olink Requirement | Metabolomics Requirement | Joint Operation Protocol |
| Blood Anticoagulant | EDTA/Heparin/Citrate | Heparin Sodium (preferred) | Uniform use of heparin sodium anticoagulant tubes |
| Plasma Volume | ≥40 µL | ≥300 µL | Aliquot into two portions, each ≥100 µL |
| Centrifugation Timeframe | ≤30 min (2–8°C) | ≤30 min (2–8°C) | Centrifuge immediately post-collection |
| Storage Temperature | -80°C | -80°C | Transport on dry ice; maintain frozen throughout |
| Tissue Weight | Lysis buffer: 0.5–1 mg/mL | ≥250 mg | Flash-freeze in liquid nitrogen; aliquot ≥100 mg per sample |
Notes
- The same biological sample should be aliquoted into two independent portions for separate Olink and metabolomics profiling.
- Blood samples are the preferred specimen type. For alternative samples including cerebrospinal fluid, urine, saliva, or tissue lysates, applicability must be pre-evaluated prior to analysis.
FAQs
What are the core advantages of integrating Olink Proteomics with Metabolomics?
Proteins and metabolites represent crucial functional molecules within organisms, exhibiting complex interconnections. By simultaneously examining changes in both protein expression and metabolite profiles, researchers gain deeper understanding of disease initiation and progression mechanisms. It enables discovery of potential therapeutic targets and novel biomarkers. This multi-omics approach reveals critical biological information undetectable through single-omics investigations alone.
How to collect samples to ensure compatibility between both technologies?
Sample collection and processing are critical for Olink technology and metabolomics analysis. To ensure compatibility between both approaches, the following points must be addressed:
- Sample types: Select appropriate types such as serum, plasma, cells, or tissues.
- Sample handling: Standardize processing protocols, including collection timing, processing methods, and storage conditions.
- Sample volume: Ensure sufficient quantity to meet both analytical requirements.
- Quality control: Perform quality checks to maintain sample integrity and purity.
Is this integrated technology suitable for specific disease research?
The combined analysis of Olink technology and metabolomics is applicable to research across various diseases, particularly in metabolic disorders and cancer investigations.
- Metabolic disorders: This approach can elucidate mechanisms and identify biomarkers for conditions such as diabetes, obesity, and cardiovascular diseases.
- Cancer research: By analyzing protein and metabolite alterations in tumor cells, novel therapeutic targets and biomarkers may be discovered, providing a basis for personalized treatment.
- Neurological disorders: The multi-omics methodology can be applied to neurodegenerative diseases like Alzheimer's disease to identify new biomarkers and therapeutic strategies.
References
- Temba GS, Pecht T, Kullaya VI et al (2025) Immune and metabolic effects of african heritage diets versus western diets in men: a randomized controlled trial. Nat Med 31(5):1698-1711. https://doi.org/10.1038/s41591-025-03602-0
- Halama A, Zaghlool S, Thareja G et al (2024) A roadmap to the molecular human linking multiomics with population traits and diabetes subtypes. Nat Commun 15(1):7111. https://doi.org/10.1038/s41467-024-51134-x
- Yamamoto Y, Shirai Y, Sonehara K et al (2025) Dissecting cross-population polygenic heterogeneity across respiratory and cardiometabolic diseases. Nat Commun 16(1):3765. https://doi.org/10.1038/s41467-025-58149-y