- Overview
- Panels List
- Applications
- Workflow
- Why Creative Proteomics
- Demo
- Sample Requirements
- Case Study
- FAQ
- Sample Submission Pack
Why Focus on the Human Proteome with Olink?
Understanding the human proteome is essential for linking genetic variation to disease biology. Olink's technology provides a uniquely powerful approach for this challenge:
- Exceptional sensitivity for human biomarkers – The dual-antibody recognition and DNA hybridization system minimizes false positives, enabling detection of proteins at femtogram levels. This makes it possible to capture low-abundance but biologically critical human proteins that often escape traditional assays.
- Scale for population-level studies – Olink can profile up to 5,343 human proteins simultaneously from just a few microliters of plasma or serum. This throughput is ideal for biobank projects, epidemiological cohorts, and longitudinal studies involving tens of thousands of participants.
- Dynamic range for diverse human samples – With a 10-log dynamic range, Olink accurately quantifies both high- and low-abundance proteins in the same assay. Minimal input requirements preserve irreplaceable clinical or biobank material.
- Integration with human genomics – Olink data pairs seamlessly with genome-wide association studies and transcriptomics, enabling the discovery of protein quantitative trait loci (pQTLs) and causal pathways in human disease.
Compared to alternatives such as SomaScan, Olink demonstrates higher target specificity and stronger phenotypic correlations—qualities that are essential for reliable human disease research.
Selecting the Right Panel for Human Proteome Studies
Mapping the human proteome requires both breadth and precision. We offer a tiered panel strategy designed specifically for human-focused research:
- Discovery Scale – Olink Explore HT
For large-scale human proteome mapping, Explore HT and Explore 3072 panels measure thousands of proteins across cardiometabolic, oncology, neurology, and inflammatory pathways. These high-plex assays are ideal for biobank cohorts, epidemiological studies, and early biomarker discovery.
- Validation Scale – Olink Target 96 and Target 48
Once candidate proteins are identified, focused panels provide hypothesis-driven validation. Covering 48–96 proteins per panel, they enable reproducible analysis of human-specific pathways such as immune response, cardiovascular health, and neurodegeneration.
- Tailored Scale – Olink Flex Panels
For research that requires maximum specificity, Olink Flex allows the design of custom human proteome panels. Investigators can target protein signatures relevant to defined disease mechanisms (e.g., inflammation in aging, cytokine storms, or immuno-oncology).
This three-level approach ensures you can move seamlessly from unbiased discovery to pathway validation to disease-specific customization—all within the context of human biology.
Here is a list of recommended panels we offer:
Table. List of Olink Panels
Additional Options
In addition to the panels above, we also offer panels for mouse-focused research:
- Target 96
- Target 48
Our team can assist in designing an integrated Olink–Human Proteome Assay strategy tailored to your project goals.
Applications of Human Proteome Assay
Our Human Proteome Assay Service is designed to answer key questions in human health and disease research. By combining high-plex discovery with targeted validation, it supports the following applications:
- Population-Scale Biomarker Discovery
Measure thousands of proteins from just a few microliters of human plasma or serum. Ideal for biobank projects, longitudinal cohorts, and epidemiological studies, this approach identifies protein signatures linked to disease risk, progression, and therapeutic response.
- Cardiovascular and Metabolic Disorders
Reveal circulating protein markers associated with heart disease, diabetes, and metabolic syndrome. Olink assays have been used in Mendelian randomization studies to connect specific proteins to clinical outcomes in large human populations.
- Oncology and Early Cancer Detection
Profile the human blood proteome to discover cancer-specific protein signatures. These insights enable early detection, patient stratification, and differentiation between tumor types—all from a low-volume blood sample.
- Neurology and Aging Research
Track proteins linked to neurodegenerative diseases, neuroinflammation, and age-related decline. Olink's sensitivity allows detection of subtle changes in low-abundance proteins critical for Alzheimer's and Parkinson's research.
- Multi-Omics Integration in Human Studies
Combine proteomic profiles with genomic and transcriptomic data to uncover protein quantitative trait loci (pQTLs) and disease-associated pathways. This systems biology approach strengthens causal inference and accelerates drug target validation.
- Complementary to Mass Spectrometry
While mass spectrometry provides broad discovery, Olink excels in sensitive quantification of low-abundance proteins in human plasma and serum. Used together, they offer unmatched depth and reliability for human proteome studies.
Workflow of Human Proteome Assay
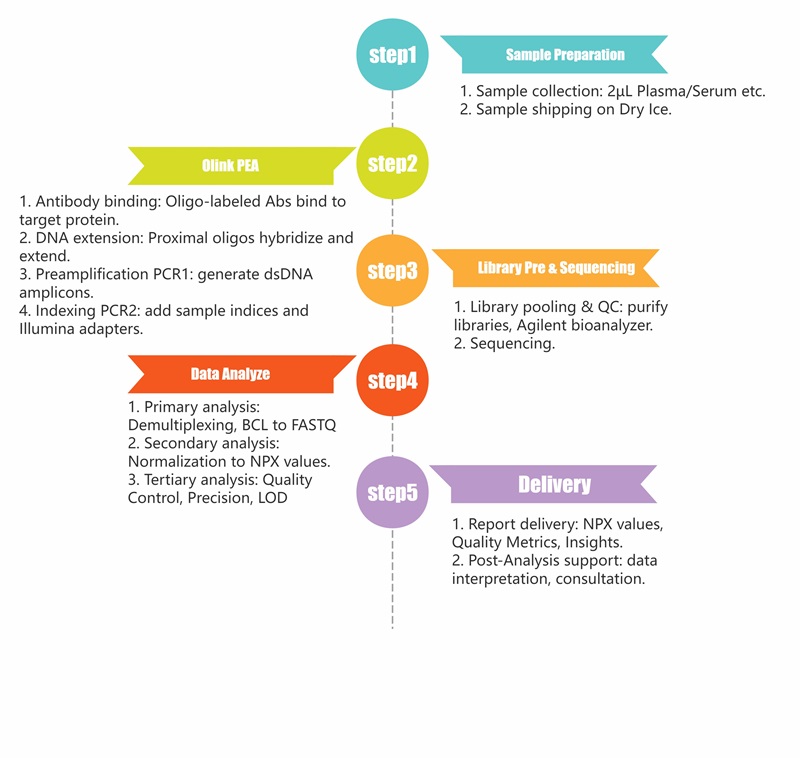 Figure 1: Workflow of Olink' s Human Proteome Assay Service.
Figure 1: Workflow of Olink' s Human Proteome Assay Service.
Why Creativ Proteomics
Studying the human proteome requires more than just a platform—it demands expertise in handling human samples, large-scale cohorts, and disease-focused questions. CPR offers a tailored solution for this challenge:
Comprehensive Human-Focused Portfolio
From broad discovery panels (Explore HT, Explore 3072) to targeted validation (Target 96/48) and custom Flex panels, we provide full coverage of the human proteome. Researchers can transition smoothly from discovery in biobank cohorts to validation in disease-specific studies.
Integration with Human Genomics and Clinical Data
Our team specializes in proteogenomic analysis, linking Olink data with GWAS, pQTL mapping, and transcriptomics. This integration uncovers causal disease pathways and strengthens translational research outcomes.
Proven Experience with Human Samples and Cohorts
We routinely process plasma and serum from large human cohorts, ensuring consistency across thousands of samples. Our protocols safeguard data quality while preserving precious biobank material.
Cutting-Edge Sensitivity for Human Biomarkers
Olink's PEA technology detects low-abundance human proteins such as cytokines and growth factors from as little as 2 µL of sample—critical for early disease detection and mechanistic research.
Rigorous Quality and Expert Support
Every project is backed by strict QC, validated workflows, and expert consultation in human study design, from disease panel selection to multi-omics integration strategies.
Together, these strengths make CPR a trusted partner for population-scale proteomics, human disease biomarker discovery, and translational research.
Demo Results: Olink Human Proteome Assay
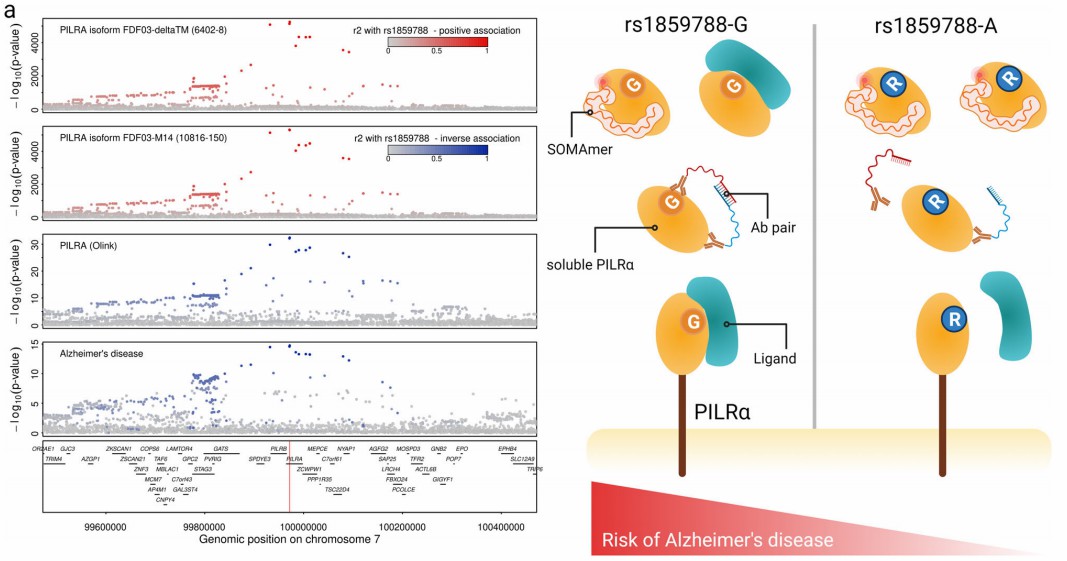 Figure 2: Regional association plots for paired immunoglobulin-like type 2 receptor alpha (PILRα) measured by SomaScan and Olink. (Pietzner, M., et al. 2021)
Figure 2: Regional association plots for paired immunoglobulin-like type 2 receptor alpha (PILRα) measured by SomaScan and Olink. (Pietzner, M., et al. 2021)
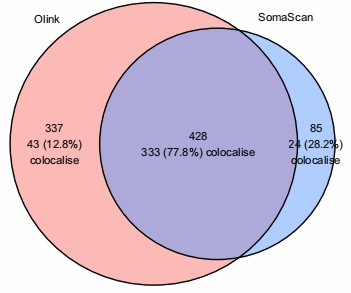 Figure 3: Number of proteins with cis-pQTLs identified in Olink and SomaScan. (Wang, B., et al. 2025)
Figure 3: Number of proteins with cis-pQTLs identified in Olink and SomaScan. (Wang, B., et al. 2025)
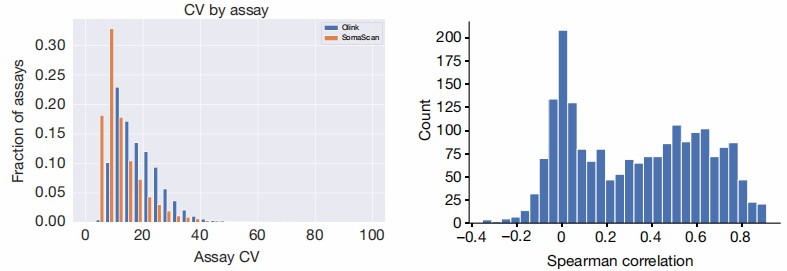 Figure 4: Protein levels measured by individual assays. (Eldjarn, G.H., et al. 2023)
Figure 4: Protein levels measured by individual assays. (Eldjarn, G.H., et al. 2023)
Sample Requirements for Human Proteome Assay
1. Supported Sample Types
- Plasma (Recommended): Olink' s protocols and validation are most extensively optimized for plasma, making it the preferred sample type.
- Serum is also accepted.
Notes
- EDTA is the strongly recommended and most validated anticoagulant.
- Other sample types like CSF, cell culture supernatants, or tissue lysates may be compatible with other Olink panels but are typically not suitable for the large-scale Human Proteome Assay without prior validation and customization.
2. Sample Collection and Processing
- Fasting State: While not always mandatory, collecting samples from participants in a consistent fasting state (e.g., overnight fast) is recommended to minimize dietary-induced variability.
- Aliquoting: Immediately aliquot the processed plasma/serum into smaller volumes after the first centrifugation. This prevents repeated freeze-thaw cycles of the main stock.
- Hemolysis, Lipemia, and Icterus:Visually inspect samples.Severely hemolyzed, lipemic, or icteric samples should be avoided or noted, as they can interfere with the assay and skew results for many proteins.
3. Sample Storage and Shipping
- Storage Temperature:Store samples at -80°Cimmediately after processing and aliquoting. Avoid using -20°C freezers for long-term storage as it is not cold enough to ensure full stability over time.
- Freeze-Thaw Cycles: Minimize freeze-thaw cycles absolutely. A maximum of 2-3 freeze-thaw cycles is generally considered acceptable, but 1 is ideal. This is because each cycle can degrade proteins and lead to diminished signal.
- Shipping: Samples must be shipped to the testing facility on dry ice to ensure they remain frozen.
4. Sample Volume and Concentration
- Minimum Volume: The minimum volume required per sample to run the Human Proteome Assay is 15 µL.
- Recommended Volume: It is highly recommended to provide 20-30 µL. This provides a sufficient volume for the assay run, any necessary repeat analysis, and potential QC re-checks without needing to thaw another aliquot.
- Concentration: The assay is designed to work with native, undiluted plasma and serum. Do not dilute your samples. The PEA technology is highly sensitive and is optimized for the complex matrix of blood.
Case Study
Title: A plasma proteomics-based candidate biomarker panel predictive of amyotrophic lateral sclerosis
Journal: Nat Med
Year: 2025
- Background
- Methods
- Results
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disorder characterized by progressive motor neuron degeneration, leading to muscle atrophy, respiratory failure, and death within 2–4 years of symptom onset. By 2040, ~400,000 people worldwide are projected to live with ALS, posing a significant healthcare burden. Current diagnosis relies on clinical symptoms and neurological exams—methods unchanged since Charcot's initial description 155 years ago—resulting in diagnostic delays of 6–18 months. The heterogeneity of ALS manifestations further complicates differentiation from other neurological diseases, delaying trial enrollment and future treatment initiation. No definitive biomarker exists, highlighting an urgent need for objective diagnostic tools to accelerate therapeutic development.
Plasma proteomic profiling was performed using the Olink Explore 3072 platform, quantifying 2,886 proteins after quality control. Key methodological steps included:
1. Platform Principle: Proximity Extension Assay (PEA) technology, where antibody-oligonucleotide probes bind target proteins, hybridize, and generate quantifiable DNA barcodes via polymerase extension.
2. Sample Processing: Plasma collected via venipuncture (heparin/EDTA tubes), centrifuged within 2 hours, aliquoted, and stored at –80°C with ≤3 freeze-thaw cycles.
3. Quality Control: Median intra- and inter-assay coefficients of variation were 9.9% and 22.3%, respectively, confirming platform reliability.
4. Normalization: Data normalized using bridging samples across cohorts (except UK Biobank data).
5. Statistical Adjustment: Analyses adjusted for age, sex, tube type, and genetic population stratification (UMAP-reduced principal components).
The results demonstrated that the Olink Explore 3072 platform reliably identified plasma proteomic signatures in ALS, as evidenced by its low median intra-assay and inter-assay coefficients of variation (9.9% and 22.3%, respectively), which confirmed the platform's robustness for large-scale biomarker discovery. Proteome-wide analysis revealed 33 differentially abundant proteins in ALS patients compared to controls (Fig. 2a and 2b), with neurofilament light chain (NEFL) showing the highest log₂ fold change (2.34, FDR-adjusted P = 2.22 × 10⁻⁸⁸), alongside novel markers such as ALDH3A1 and MEGF10; this differential abundance was replicated in an independent cohort with high concordance (R = 0.83, P = 1.80 × 10⁻⁹), as visualized in the volcano plots and scatter plot.
Furthermore, comparisons with orthogonal methods validated Olink's accuracy, as shown in Extended Data Fig. 1, where protein measurements from Olink strongly correlated with ELISA and ProQuantum assays (e.g., Pearson correlations for key proteins like NEFL and CSRP3), with Bland-Altman plots indicating minimal bias.
Additionally, Extended Data Fig. 2 highlighted Olink's broad coverage and consistency with the SomaScan platform in overlapping proteins. These Olink-based findings formed the foundation for the machine learning model, which achieved an AUC of 98.3% in diagnosing ALS, underscoring the technology's utility for high-precision proteomic profiling in neurodegenerative diseases.
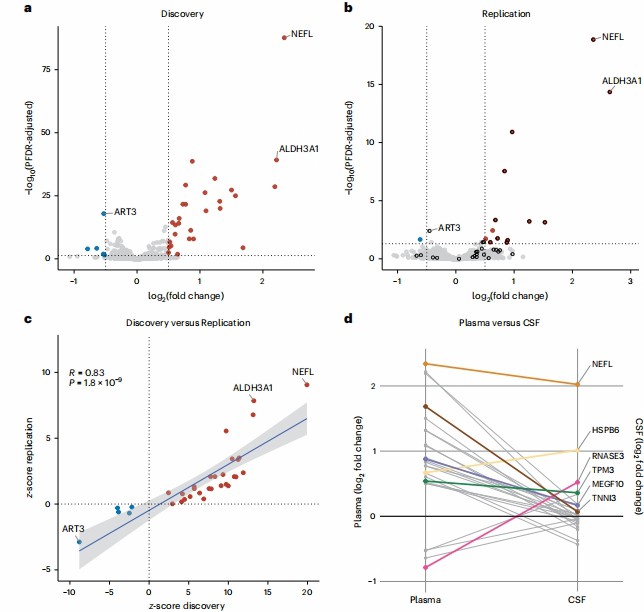 Figure 5: Differential abundance of plasma proteins in patients diagnosed with ALS compared with control individuals. (Chia, R., et al. 2025)
Figure 5: Differential abundance of plasma proteins in patients diagnosed with ALS compared with control individuals. (Chia, R., et al. 2025)
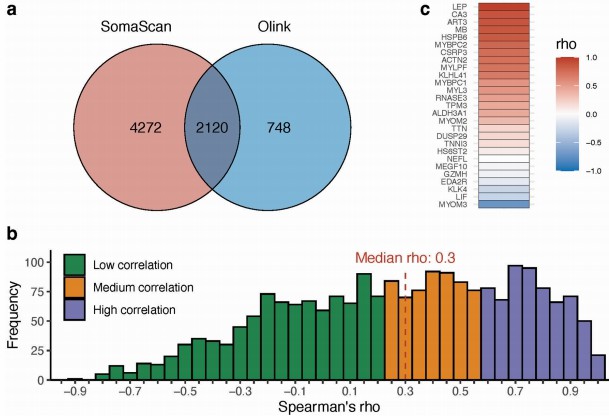 Figure 6: Comparison between SomaScan 7k and Olink 3072 Explore in BLSA samples. (Chia, R., et al. 2025)
Figure 6: Comparison between SomaScan 7k and Olink 3072 Explore in BLSA samples. (Chia, R., et al. 2025)
FAQs
How Does Olink Compare to Other Proteomic Technologies (e.g., Mass Spectrometry, SomaScan)?
| Feature | Olink PEA | Mass Spectrometry | SomaScan |
| Principle | Immunoassay + NGS | Measures peptide mass | Aptamer binding + fluorescence |
| Multiplexing | High (Excellent) | Medium to High (can be limited) | High |
| Sensitivity | Very High | Moderate (challenge for low-abundance) | High |
| Specificity | Very High (Dual Ab) | High (can suffer interference) | High (potential off-target) |
| Sample Volume | Very Low (e.g., 2 µL) | Generally higher | Low (but often more than Olink) |
| Primary Readout | Relative Quantification (NPX) | Can provide Absolute Quantification | Relative Quantification |
What are the Main Applications?
Olink assays are widely used in:
- Biomarker Discovery: Identifying protein biomarkers for early disease diagnosis, prognosis, and monitoring treatment response in conditions like Alzheimer' s disease and various cancers.
- Drug Development: Aiding in target identification, patient stratification, and assessing drug efficacy and toxicity.
- Large-Scale Population Studies: Enabling large cohort studies to understand associations between the proteome, genetics, and diseases. A prominent example is its use in the UK Biobank project.
- Basic Mechanistic Research: Investigating disease mechanisms and biological pathways.
References
- Pietzner, M., Wheeler, E., Carrasco-Zanini, J. et al. Synergistic insights into human health from aptamer- and antibody-based proteomic profiling. Nat Commun 12, 6822 (2021).
- Wang, B., Pozarickij, A., Mazidi, M. et al. Comparative studies of 2168 plasma proteins measured by two affinity-based platforms in 4000 Chinese adults. Nat Commun 16, 1869 (2025).
- Eldjarn, G.H., Ferkingstad, E., Lund, S.H. et al. Large-scale plasma proteomics comparisons through genetics and disease associations. Nature 622, 348–358 (2023).
- Chia, R., Moaddel, R., Kwan, J.Y. et al. A plasma proteomics-based candidate biomarker panel predictive of amyotrophic lateral sclerosis. Nat Med (2025).