- Overview
- Panels List
- Applications
- Workflow
- Why Creative Proteomics
- Demo
- Sample Requirements
- FAQ
- Sample Submission Pack
Why Integrate Olink Proteomics with mRNA-seq?
The integration of Olink proteomics and mRNA-seq leverages two complementary logics to maximize biological insight. Vertical validation uses RNA-seq to identify candidate genes at the transcriptional level, followed by Olink to confirm corresponding protein-level changes. Thereby it verifies functional biomarker relevance and filters false positives from transcriptional noise. Horizontal complementarity exploits RNA-seq' s capacity to reveal upstream transcriptional regulators. While Olink detects downstream effector proteins, uncovering regulatory cascades. Transcriptional drivers and functional proteomic outputs may diverge due to post-translational modifications or secretion dynamics. This dual-mode integration enables robust biomarker discovery and mechanistic dissection of disease heterogeneity.
Olink Technology
Proximity Extension Assay (PEA)
1) Antibody pairs conjugate to DNA oligonucleotides.
2) Antibody pairs bind to target protein.
3) Upon co-localization, the DNA strands hybridize and extend into amplifiable templates.
4) Quantifiable DNA signals via qPCR or NGS.
mRNA-seq
1) Total RNA extraction.
2) mRNA enrichment (poly-A selection/rRNA depletion).
3) cDNA synthesis.
4) NGS library construction.
5) High-throughput sequencing.
6) Alignment to a reference genome.
7) Quantification of gene expression, differential expression (DEGs), splicing variants, and novel transcripts.
Advantages
- Minimal Sample Requirements: Olink uses ≤6 µL of plasma; mRNA-seq requires µg-level total RNA.
- Ultra-High Sensitivity: Olink detects low-abundance proteins missed by mass spectrometry.
- Multi-Omics Depth: Integrate genetic variants, transcriptomics, and proteomics for mechanistic insights.
- Scalability: Olink Explore panels support 1,536–3,072 proteins per run via NGS.
Olink & mRNA-seq Platform: Selecting the Right Panel
The selected panels are chosen for their collective ability to cover diverse biological pathways and research areas. From foundational physiological processes to specific immune responses, neurological functions, organ health, and various oncology contexts, this broad coverage provides complementary protein biomarker data that can be powerfully correlated with transcriptomic profiles obtained via mRNA-Seq.
Including both broad, multiplex panels (Explore) and focused, targeted panels (Target 96/48, Customized Flex) allows researchers to tailor the protein analysis to their specific hypotheses. Leveraging the technology's capability can uncover novel biological insights and validate connections between gene expression and corresponding protein levels and activities.
Here is a list of recommended panels we offer:
Table. List of Olink Panels
Applications
Olink and RNA-seq integration transforms biological research—from dissecting cellular heterogeneity to validating cross-species mechanisms—by bridging gene-to-protein dynamics with high sensitivity and scalability.
Decoding Cellular Heterogeneity and Functional States
- Cell Subtype Characterization: Combined scRNA-seq (single-cell resolution) + Olink proteomics (e.g., Target 96 Inflammation Panel) to profile cell populations.
- Environmental Response Dynamics: Longitudinal Olink (plasma proteomics) + RNA-seq tracked molecular responses to environmental triggers.
Biomarker Discovery and Validation
- Target Prioritization: RNA-seq nominates candidate genes; Olink confirms protein-level changes, filtering false positive.
- Multi-Omics Biomarker Panels
Molecular Response Profiling
- Stimulus-Sensitive Pathways: 1) RNA-seq revealed obesity-induced transcriptional regulators, Olink detected downstream effector proteins in mechanically stressed tissues. 2) Olink quantified secreted proteins in response to TNF-α stimulation; RNA-seq linked these to glycolytic pathway upregulation.
- Immune Activation Signatures: Integrated Olink + CyTOF + PBMC RNA-seq decoded Omicron-specific protein signatures post-vaccination.
Cross-Species Biological Conservation
- Olink Target 48 Mouse Cytokine Panel + RNA-seq profiled conserved immune proteins in model organisms under mechanical stress.
Workflow of Olink & mRNA-seq
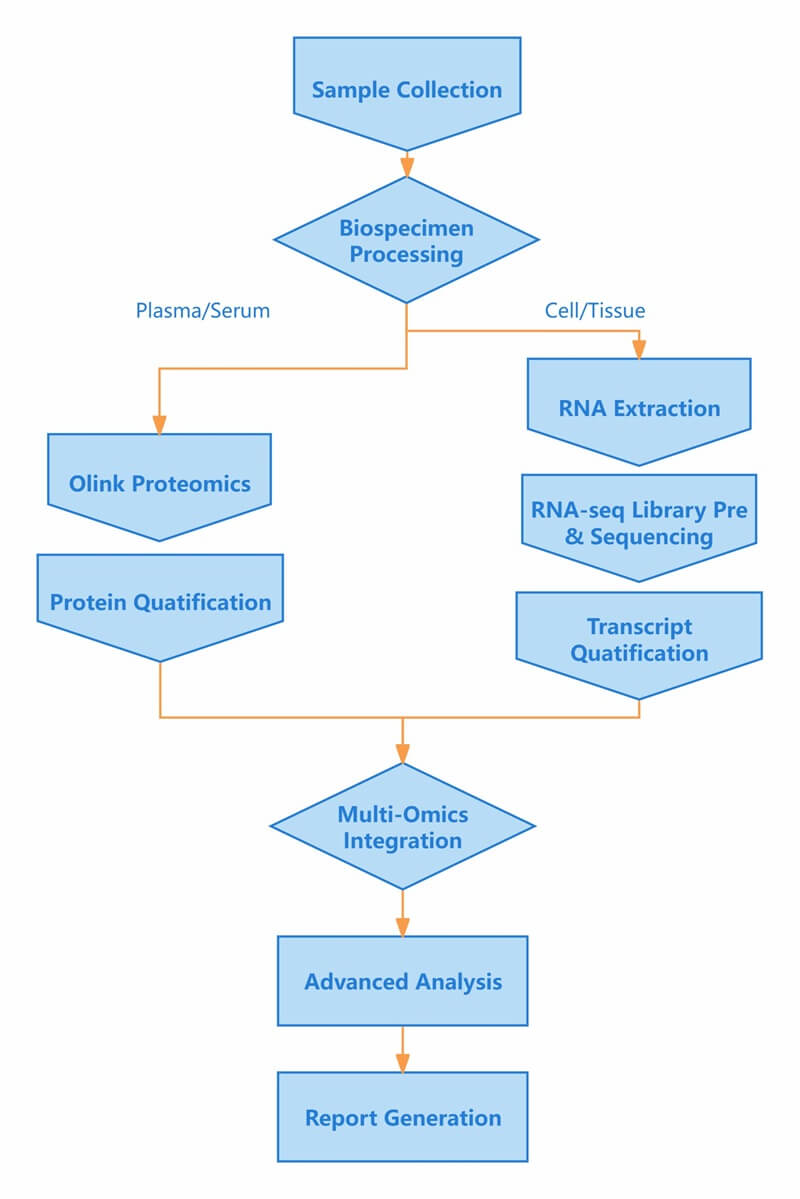 Figure 1: Workflow of integrated Olink proteomics and mRNA-seq.
Figure 1: Workflow of integrated Olink proteomics and mRNA-seq.
Why Creativ Proteomics
Cutting-Edge Technical Platforms & Certifications
- Olink Platform Mastery: Proficiency in Olink' s PEA technology, which enables ultrasensitive detection of low-abundance proteins in complex samples. This includes validated workflows for panels like Explore 1536 or Target 96/384.
- NGS and mRNA-seq Capabilities: Expertise in full-length mRNA sequencing with optimizations for single-cell or bulk RNA-seq, ensuring high-resolution transcriptome data.
- Certifications: Official Olink accreditation and NGS platform certifications to guarantee protocol adherence and reproducibility.
Integrated Multi-Omics Analysis Expertise
- Cross-Platform Data Integration: Ability to reconcile discordant mRNA-protein correlations.
- Biological Interpretation Skills: Experience in linking transcriptional regulators to downstream protein effects. Tools for pathway enrichment and trajectory analysis are essential.
Stringent Quality Control (QC) and Low-Input Protocols
- Sample Handling: Optimized workflows for minimal sample volumes. Expertise in handling challenging samples.
- QC Metrics: Implementation of plate/background normalization (Olink), detection thresholds, and RNA integrity checks.
End-to-End Service Design & Support
- Consultative Project Design: Guidance on panel selection, time-point sampling for dynamic processes, and cohort size optimization.
- Customization: Flexibility to tailor workflows, e.g., co-profiling intracellular proteins + mRNA via techniques or integrating microbiome data with host proteomics.
- Turnkey Solutions: From wet-lab processing to cloud-based bioinformatics.
Demo Results
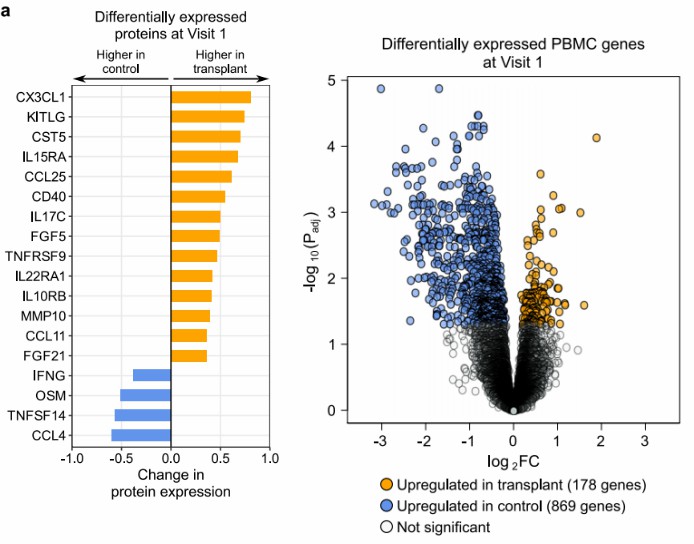 Figure 2: SOT recipients demonstrate persistently elevated serum chemokines, reduced IFN-gamma, and enhanced innate immune gene expression in PBMCs throughout COVID-19 hospitalization. (Pickering, H., et al. 2025)
Figure 2: SOT recipients demonstrate persistently elevated serum chemokines, reduced IFN-gamma, and enhanced innate immune gene expression in PBMCs throughout COVID-19 hospitalization. (Pickering, H., et al. 2025)
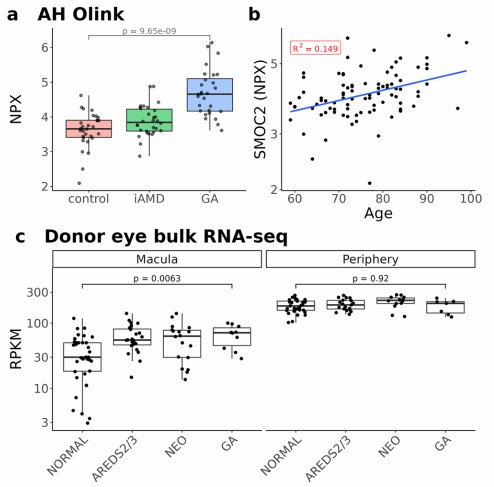 Figure 3: SMOC2 protein levels are significantly elevated in the aqueous humor of GA patients, show a positive correlation with age, and demonstrate upregulated expression in the retinal tissue of AMD donor eyes. (Huang, K., et al. 2025)
Figure 3: SMOC2 protein levels are significantly elevated in the aqueous humor of GA patients, show a positive correlation with age, and demonstrate upregulated expression in the retinal tissue of AMD donor eyes. (Huang, K., et al. 2025)
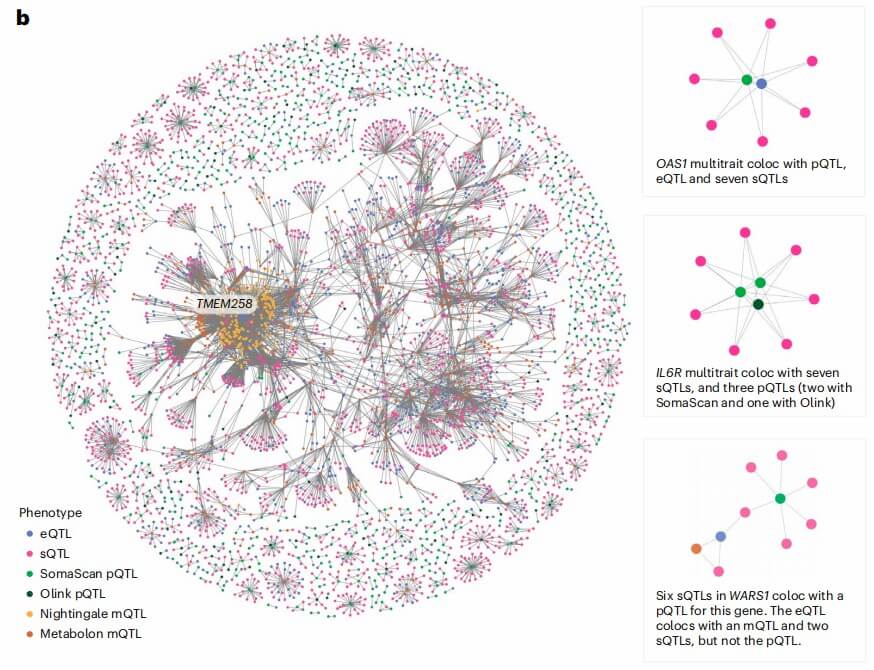 Figure 4: etwork graph of all pairwise colocalization results. (Tokolyi, A., et al. 2025)
Figure 4: etwork graph of all pairwise colocalization results. (Tokolyi, A., et al. 2025)
Sample Requirements
| Step | Olink Requirement | RNA-seq Requirement |
| Volume | 1-6 µL per panel | ≥100 ng total RNA |
| Sample Types | Serum, plasma, CSF, tissue lysate | Cells, tissue, blood |
| Storage Temperature | -80°C (for long term), avoid freeze-thaw cycles | 4°C refrigeration (for short term); –80°C freezing (for long term) |
Notes
- RIN ≥7 for bulk RNA-seq; live cells for scRNA-seq.
- Compatibility: Use matched samples (e.g., same plasma aliquot for Olink, PBMCs for RNA-seq).
- Preservation: Snap-freeze tissues in liquid N₂; use EDTA plasma for Olink to avoid platelet contamination.
FAQs
What is the core advantage of combining Olink Proteomics with RNA-seq?
The integration provides multi-dimensional insights into disease mechanisms. Olink detects low-abundance proteins in minimal sample volumes (1–6 µL) with high sensitivity (fg/mL range). RNA-seq reveals transcriptome-wide gene expression, splicing variants, and non-coding RNAs. Together, they bridge the gap between mRNA expression and actual protein levels, overcoming limitations of transcript-protein correlation (especially for low-abundance proteins). This synergy identifies novel biomarkers and disease endotypes.
How are Olink and RNA-seq integrated in study design?
A typical workflow includes:
Sample Processing: Use the same biospecimen (e.g., serum, tissue lysate) for both assays. Olink supports diverse samples (serum, plasma, cell supernatants).
Parallel Analysis: Olink panels quantify 96–3,072 proteins and RNA-seq (bulk or single-cell) profiles transcriptomes.
Data Integration: Cross-validate findings by identifying overlapping pathways and using transcriptome data to prioritize proteins for Olink validation.
Functional Validation: Immunofluorescence, Luminex, or metabolic assays confirm key targets.
Can Olink replace RNA-seq for biomarker discovery?
No. The technologies are complementary in biological research. RNA-seq provides a comprehensive, untargeted view of the transcriptome, enabling genome-wide detection of all RNA transcripts. This makes it indispensable for initial biomarker screening and hypothesis generation. RNA-seq identifies gene-level alterations, Olink validates functional protein-level changes, bridging the critical gap between mRNA expression and actual protein abundance. Thus, integrating both methods resolves transcript-protein discordance, enabling end-to-end biomarker validation. Researchers typically use RNA-seq for broad discovery and Olink for focused verification, maximizing cost-efficiency and biological insights.
What are key technical considerations for integration?
Based on the technical considerations for integrating Olink and RNA-seq workflows, key factors to address include sample compatibility, cost efficiency, data harmonization, and scalability, ensuring robust multi-omics insights. Sample quality is paramount. For cost efficiency, researchers often prioritize RNA-seq for broad, untargeted screening to identify candidate biomarkers or pathways, followed by Olink' s focused protein validation. Data integration leverages specialized tools. Scalability is enhanced by Olink' s high-throughput panels, which align with RNA-seq batch processing for large cohorts.
References
- Pickering, H., Schaenman, J., Phan, H.V. et al. Host-microbe multiomic profiling identifies distinct COVID-19 immune dysregulation in solid organ transplant recipients. Nat Commun 16, 586 (2025). https://doi.org/10.1038/s41467-025-55823-z
- Huang, K., Schofield, C., Nguy, T. et al. Proteomics approach identifies aqueous humor biomarkers in retinal diseases. Commun Med 5, 134 (2025). https://doi.org/10.1038/s43856-025-00862-2
- Tokolyi, A., Persyn, E., Nath, A.P. et al. The contribution of genetic determinants of blood gene expression and splicing to molecular phenotypes and health outcomes. Nat Genet 57, 616–625 (2025). https://doi.org/10.1038/s41588-025-02096-3