- Overview
- Panels List
- Applications
- Workflow
- Why Creative Proteomics
- Demo
- Sample Requirements
- Case Study
- FAQ
- Sample Submission Pack
Why Use Olink for Disease Classification?
Olink excels in Protein Biomarker Profiling by offering unparalleled scalability (>5,400 proteins simultaneously), minimal sample requirements (ideal for longitudinal/biobank studies), and rigor sufficient for experimental validation. Its PEA technology avoids the limitations of mass spectrometry (e.g., loss of low-abundance proteins) and ELISA (low throughput). By providing reproducible, high-quality data, Olink enables researchers to transition from exploratory research to practical applications.
Selecting the Right Panel for Disease Classification
The choice of panel should be driven by the stage and specific focus of your research. The selection of the panels is based on the following criteria:
- For Broad, Unbiased Discovery and Hypothesis Generation
To perform an untargeted, hypothesis-free screen across the proteome. This is the first step in classifying diseases with unknown or complex biomarkers. Explore 3072 and Explore 1536 are ideal for exploratory research in oncology, neurology, or cardiometabolic diseases where pathways are interconnected.
- For Focused, Biology-Driven Investigation
To perform a deep, quantitative analysis within a specific biological pathway or disease area. Olink Target 96 Panels is for validating discoveries or testing specific hypotheses. To analyze a core set of well-established biomarkers with high efficiency, Olink Target 48 Panels are often for validation or large-scale screening studies.
- For Tailored, Hypothesis-Led Validation
To create a bespoke panel that perfectly addresses a unique research question or validates a specific signature from a discovery-phase experiment.
Here is a list of recommended panels we offer:
Table. List of Olink Panels
Additional Options
In addition to the panels above, we also offer the following panels:
- Explore 384
- Target 96
- Custom Olink Flex systems
Our team can assist in designing an integrated Olink–Disease Classification strategy tailored to your project goals.
Applications of Disease Classification
- Biomarker Discovery for Early Disease Detection
Olink's Proximity Extension Assay (PEA) technology enables highly sensitive and specific quantification of proteins, facilitating the identification of novel biomarkers for early disease detection. The platform's sensitivity allows detection of protein expression changes before clinical manifestations become apparent, enabling earlier intervention opportunities. The simultaneous measurement of multiple biomarkers improves classification accuracy beyond what single-marker analyses can achieve.
- Disease Mechanism and Pathway Characterization
Olink proteomics provides insights into disease mechanisms by revealing protein expression patterns associated with specific biological pathways. By analyzing protein co-expression networks and conducting pathway enrichment analyses, researchers can identify activated or suppressed signaling pathways in different disease states. The technology's ability to generate protein expression data compatible with multi-omics integration further enhances mechanistic insights.
- Monitoring Disease Progression and Treatment Response
Olink proteomics enables dynamic assessment of biological changes throughout disease courses and following interventions. The platform's quantitative precision (using Normalized Protein eXpression or NPX values) allows detection of subtle protein expression changes over time, providing a molecular timeline of disease evolution.
- Multi-Disease Protein Signature Development
Olink's high-throughput capabilities facilitate the development of comprehensive protein signatures that classify and distinguish multiple disease states simultaneously. By analyzing thousands of samples, researchers can identify protein patterns characteristic of various pathological conditions, creating classification systems that transcend traditional disease boundaries.
Workflow of Disease Classification
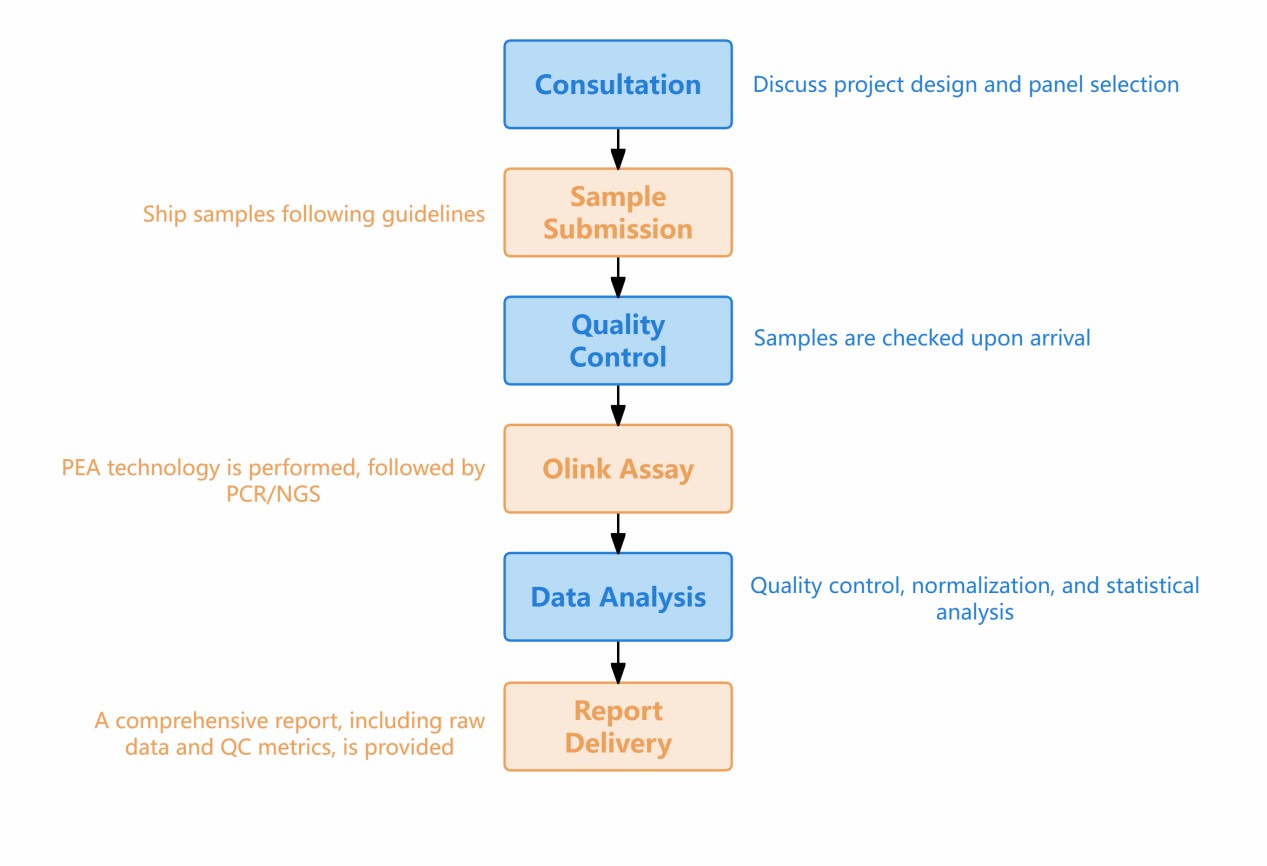 Figure 1: Workflow of Olink's Disease Classification Service.
Figure 1: Workflow of Olink's Disease Classification Service.
Why Creative Proteomics
Unmatched Analytical Performance for Precise Molecular Phenotyping
Our Olink platform provides exceptional specificity and sensitivity, effectively circumventing the cross-reactivity issues often encountered in traditional multiplex immunoassays. For disease classification, this analytical robustness ensures that the protein signatures used to distinguish diseases are accurate and reliable, minimizing false positives/negatives. This high sensitivity is particularly critical for early-stage disease detection, where biomarker concentrations are minimal.
Comprehensive, Disease-Focused Panel Design for Targeted Hypothesis Testing
Unlike broad, untargeted discovery platforms, Olink offers curated panels specifically designed for disease classification research. Our Olink Target series provides pre-configured panels focusing on 11 key disease areas, including Cardiovascular, Neurology, Oncology, Inflammation, and Metabolism. Each panel contains 92 carefully selected protein biomarkers (plus controls) relevant to the biological pathways of those diseases. This targeted approach is highly efficient.
High-Plex Profiling from Minimal Sample to Reveal Complex Biomarker Signatures
A key advantage for classification is the ability to move beyond single biomarkers to complex, multi-protein signatures. Diseases are rarely governed by a single protein; instead, they manifest as alterations in intricate networks. Our Olink technology enables high-plex protein profiling, simultaneously quantifying dozens to thousands of proteins from a single, minute sample aliquot.
Robust Data Quality and Biological Relevance for Validated Classification Models
The protein signatures we help discover for disease classification are more likely to be causally linked to the underlying disease etiology. This conviction is rooted in the core technological advantages of our platform, which are essential for developing classifiers that are both robust and reproducible across diverse patient cohorts.
Integrated Data Analysis for Biomarker Discovery and Interpretation
Our service extends beyond raw data delivery to include powerful software for interpretation. Our team helps users analyze, visualize, and derive meaning from their complex proteomic data. For disease classification, researchers can not only build a predictive model but also gain insights into the disturbed biological mechanisms that differentiate one disease class from another, understanding to the classification output.
Demo Results: Olink-Disease Classification
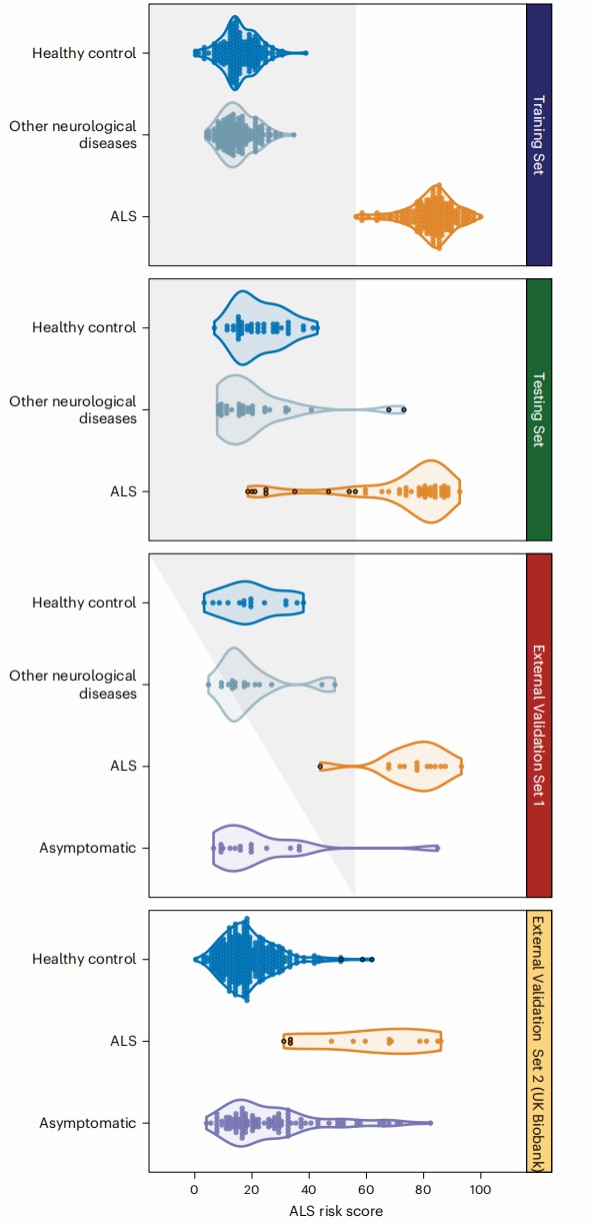 Figure 2: Classification of individual samples using the ALS risk scores generated by the random forest model using the 20 features. (Chia, R., et al. 2025)
Figure 2: Classification of individual samples using the ALS risk scores generated by the random forest model using the 20 features. (Chia, R., et al. 2025)
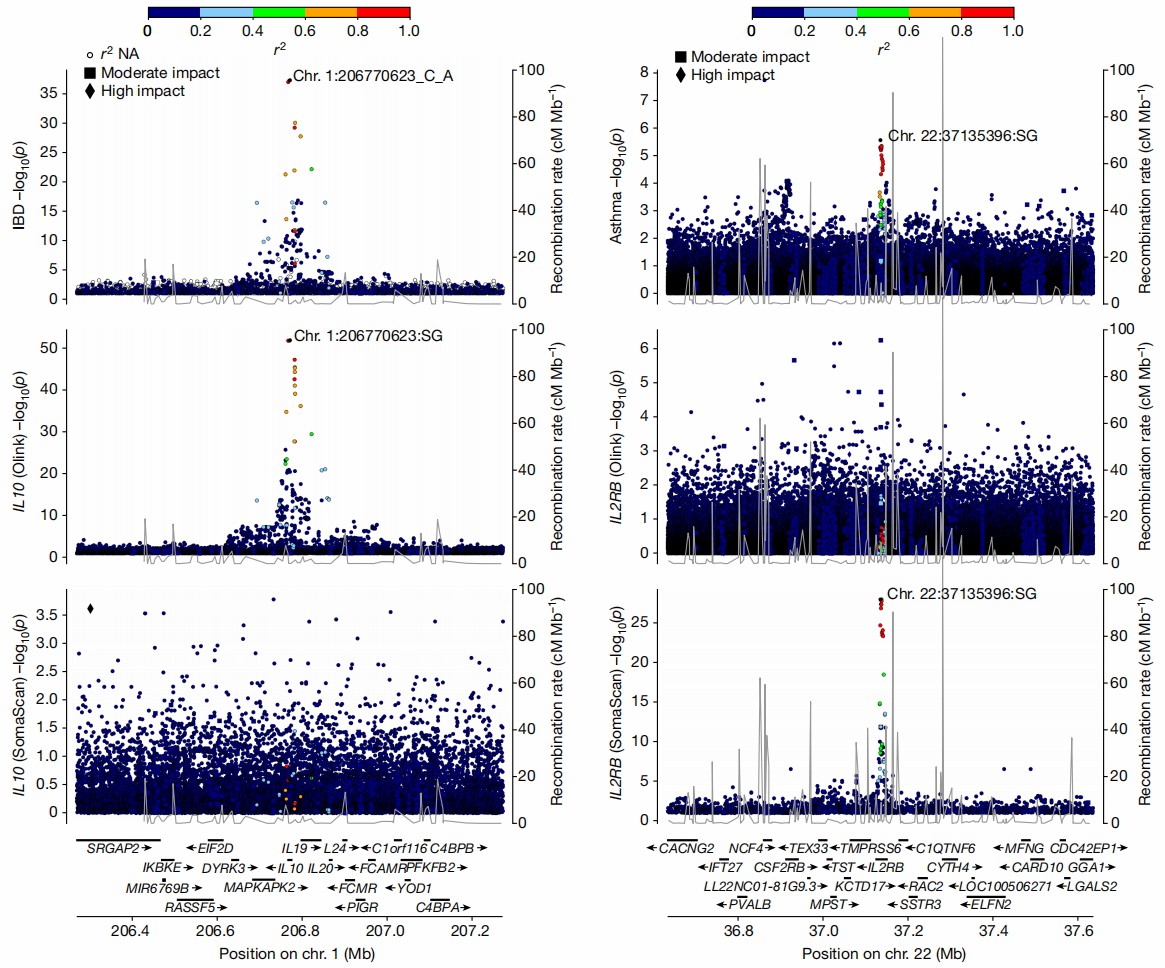 Figure 3: pQTLs that are detected on one platform only and their relationship with disease-associated variants(middle: Olink; bottom: SomaScan). (Eldjarn, G.H., et al. 2023)
Figure 3: pQTLs that are detected on one platform only and their relationship with disease-associated variants(middle: Olink; bottom: SomaScan). (Eldjarn, G.H., et al. 2023)
Sample Requirements for Olink-Disease Classification
1. Sample Types and Volumes
- Plasma/Serum (Recommended): Minimum 40-50 µL per sample is required.
- Other validated types: Cerebrospinal fluid, aqueous humor - 50 µL.
- Non-standard Sample Types (require pre-experiment validation): Cell culture supernatants, tissue lysates, cell lysates and exosome preparations.
2. Sample Storage and Shipping
- Storage temperature: -80°C.
- Shipping condition: On dry ice (samples must remain frozen upon arrival)
- Sample containers: Use temperature-resistant, non-protein-binding plasticware.
- Recommended: 96-well PCR plates with complete skirts and quality sealing membranes.
Case Study

Proteomic signatures improve risk prediction for common and rare diseases
Journal: Nat Med
Year: 2024
- Background
- Methods
- Results
For many diseases there are delays in diagnosis due to a lack of objective biomarkers for disease onset. Here, in 41,931 individuals from the United Kingdom Biobank Pharma Proteomics Project, researchers integrated measurements of approximately 3,000 plasma proteins with clinical information to derive sparse prediction models for the 10-year incidence of 218 common and rare diseases (ranging from 81 to 6,038 cases). They then compared prediction models developed using proteomic data with models developed using either basic clinical information alone or clinical information combined with data from 37 clinical assays. The predictive performance of sparse models including as few as 5 to 20 proteins was superior to models using basic clinical information for 67 pathologically diverse diseases, highlighting the potential of proteomic signatures to address diagnostic challenges in precision medicine.
Proteomic profiling was performed in EDTA-plasma samples from approximately 54,000 UK Biobank participants as part of the UK Biobank Pharma Proteomics Project (UKB-PPP), using the Olink Explore 1536 and Explore Expansion platforms. These platforms targeted 2,923 unique proteins by 2,941 assays, based on proximity extension assays where pairs of antibodies conjugated to oligonucleotides bind to target proteins, enabling hybridization, amplification, and relative quantification through next-generation sequencing. Quality control involved internal and external controls, with normalized protein expression (NPX) values generated after log2 transformation and normalization. Missing values were imputed using the missForest R package, and assays were grouped into panels such as inflammation, oncology, cardiometabolic, and neurology, ensuring robust data for downstream analysis.
The results demonstrated that sparse protein signatures derived from Olink proteomic data significantly enhanced disease prediction across diverse conditions. For example, as illustrated in Fig. 2, the addition of 5 to 20 proteins to clinical models improved the concordance index (C-index) for 67 diseases, with a median increase of 0.07 (range: 0.02–0.31), and diseases like multiple myeloma showed a delta C-index of 0.25, driven by proteins such as FCRLB and TNFRSF17, which were specifically expressed in plasma cells according to single-cell RNA sequencing data. Furthermore, protein-based models outperformed those incorporating clinical assays for 52 diseases, as seen in Fig. 3, where the likelihood ratios for proteins were higher, emphasizing the superiority of Olink-derived signatures in predicting conditions such as non-Hodgkin lymphoma and pulmonary fibrosis. External validation in the EPIC-Norfolk study confirmed the generalizability of these models, with comparable C-indexes, reinforcing the clinical utility of Olink-based proteomic signatures for early risk stratification.
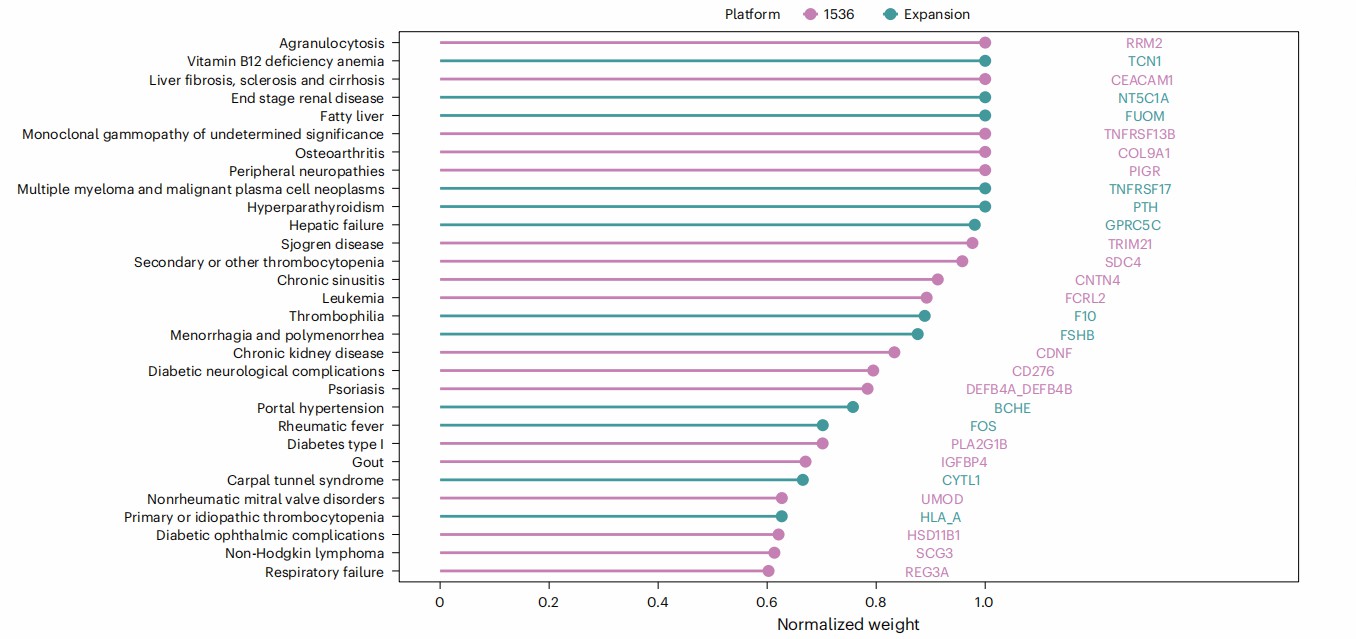 Figure 4: Disease specificity of predictor proteins. (Carrasco-Zanini, J., et al. 2024)
Figure 4: Disease specificity of predictor proteins. (Carrasco-Zanini, J., et al. 2024)
FAQs
What diseases are most suited to Olink-based classification?
Olink excels in diseases with:
- Systemic protein signatures: Dementia, cancer, cardiovascular diseases.
- Low-abundance biomarkers: Neurological disorders (where blood-brain barrier penetration is limited).
What are common pitfalls in Olink studies, and how can they be avoided?
Low detection rates can be caused by poor sample quality (e.g., hemolysis, repeated freeze-thaws). To improve this, we recommend you use EDTA plasma/serum, avoid hemolysis, and store samples at –80°C.
Batch effects occur when samples are grouped improperly. Solution is to randomize samples across plates and apply post-hoc normalization (e.g., intensity normalization for randomized studies).
High background noise is often due to sample contaminants (e.g., lipids). But pre-screen samples and optimize dilution can overcome this problem.
Can I compare results from different Olink panels?
While absolute NPX values may differ between panels, the relative expression ratios for a given protein across samples are expected to be consistent.
References
- Chia, R., Moaddel, R., Kwan, J.Y. et al. A plasma proteomics-based candidate biomarker panel predictive of amyotrophic lateral sclerosis. Nat Med (2025).
- Eldjarn, G.H., Ferkingstad, E., Lund, S.H. et al. Large-scale plasma proteomics comparisons through genetics and disease associations. Nature 622, 348–358 (2023).
- Carrasco-Zanini, J., Pietzner, M., Davitte, J. et al. Proteomic signatures improve risk prediction for common and rare diseases. Nat Med 30, 2489–2498 (2024).