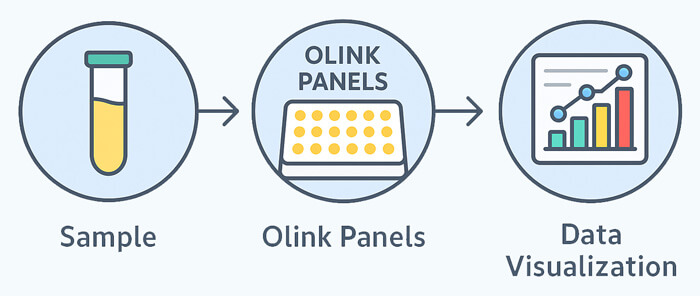
What is Olink Proteomics Panel?
Olink targeted proteomics uses a dual-antibody approach to pinpoint proteins of interest. When both antibodies bind, short DNA tags attached to each antibody pair up, are amplified, and converted into measurable signals. This mechanism enables high-throughput quantification of proteins in a variety of biological fluids — including serum, plasma, urine, and cerebrospinal fluid — from just tiny volumes.
Today, Olink proteomics technology is applied across a wide spectrum of life science research. It supports clinical research studies, drug development, diagnostic assay design, and even drug target identification. Academic scientists also rely on it to probe fundamental biological processes.
By providing a robust and ultra-sensitive proteomics platform, Olink has become a cornerstone for precision proteomics, powering research across academia and industry. Publications citing Olink-based studies continue to rise rapidly each year, underscoring its growing influence and trusted reputation.
How Does Olink Proteomics Work
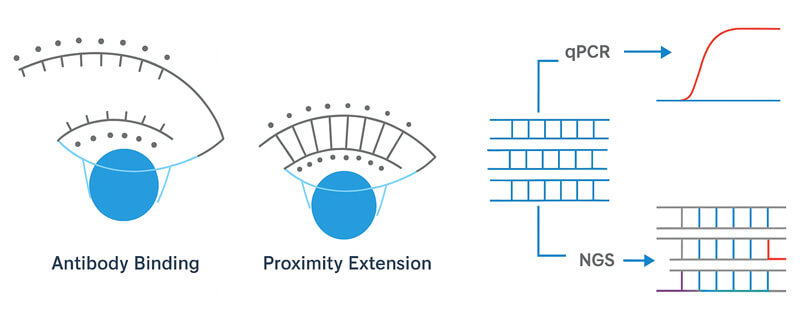
Proximity Extension Assay (PEA) is one of the most innovative advances in targeted proteomics. It combines antibody-based protein detection with oligonucleotide amplification to deliver high specificity, high sensitivity, and true high-throughput performance.
The principle is simple but powerful. Two antibodies, each tagged with a unique DNA probe, bind to the same target protein. Only when both antibodies are correctly attached do the DNA tags come close enough for their 5-base ends to align. This triggers an enzymatic extension, forming a double-stranded DNA template.
That template is then amplified and quantified using qPCR or next-generation sequencing (NGS). The resulting nucleotide signal directly corresponds to the amount of protein present, providing precise quantitative data across hundreds or thousands of targets in a single run.
By fusing immunoassay precision with molecular readouts, PEA underpins Olink's reputation for ultra-sensitive, multiplex protein analysis—an essential tool for biomarker discovery and translational research.
Key Features of Olink Proteomics Technology
Olink proteomics has become a trusted platform for biomarker discovery because of its unique blend of sensitivity, scalability, and flexibility.
High throughput
Detect 48 to 3,072 protein biomarkers per run, enabling rapid screening of large sample sets.
Minimal sample input
Requires just 1–6 μL of sample, making it ideal for scarce or precious materials.
Targeted design
Panels cover only biologically relevant proteins, spanning nearly all human signalling pathways.
Exceptional sensitivity
Reaches femtogram per millilitre (fg/mL) levels, capturing even trace proteins.
Wide dynamic range
Covers 10 orders of magnitude, accurately quantifying high-, medium-, and low-abundance proteins.
Automated workflow
Automated handling reduces manual errors, ensuring consistency and reproducibility.
Flexible detection methods
Data can be generated via qPCR or next-generation sequencing (NGS) platforms.
Liquid biopsy ready
Compatible with diverse biofluids and especially effective for detecting low-abundance proteins in serum, plasma, and CSF.
By combining these features, Olink delivers robust, publication-quality proteomics data to researchers working in drug discovery, translational studies, and precision medicine research.
Our Olink Proteomics Panel Portfolio
| Product Series | Panel Name | Quantification Type | Detection Platform | Samples per Run | Protein Targets | Key Capabilities | Applications |
| Olink Explore | 3072 | Relative | NGS | 88 | 2926 | ✅ Biomarker discovery ✅ Mechanism studies ✅ Drug target exploration |
Inflammation, oncology, neurology, cardiometabolic, organ damage, etc. |
| Olink Explore | 384 | Relative | NGS | 88 | 368/1472 | Same as above | Same as above |
|
|
1536 | Relative | NGS | 88 | 1472 | Same as above | Same as above |
|
|
HT (High-Throughput) | Relative | NGS | 172 | 5300+ | Same as above | Same as above |
| Olink Target | Target 96 | Relative | Signature Q100 | 88 | 92 | ✅ Biomarker validation ✅ Mechanistic studies |
Inflammation, oncology, neurology, cardiometabolic, immune response |
|
|
Target 48 | Absolute/Relative | Signature Q100 | 40 | 45 | ✅ Biomarker validation ✅ Target research |
Cytokine and immune studies |
|
|
Target 48 Mouse | Absolute/Relative | Signature Q100 | 40 | 43 | ✅ Preclinical biomarker research | Mouse-specific cytokine research |
| Olink Flex | Customizable Flex | Absolute/Relative | Signature Q100 | 40 | 21–30 (custom) | ✅ Custom biomarker selection ✅ Drug target research |
Tailored research projects, exploratory studies |
| Olink Focus | Focus Panels | Absolute/Relative | Signature Q100 | 144 | ≤21 | ✅ Biomarker validation | Inflammation, oncology, neurology, cardiometabolic, immune response |
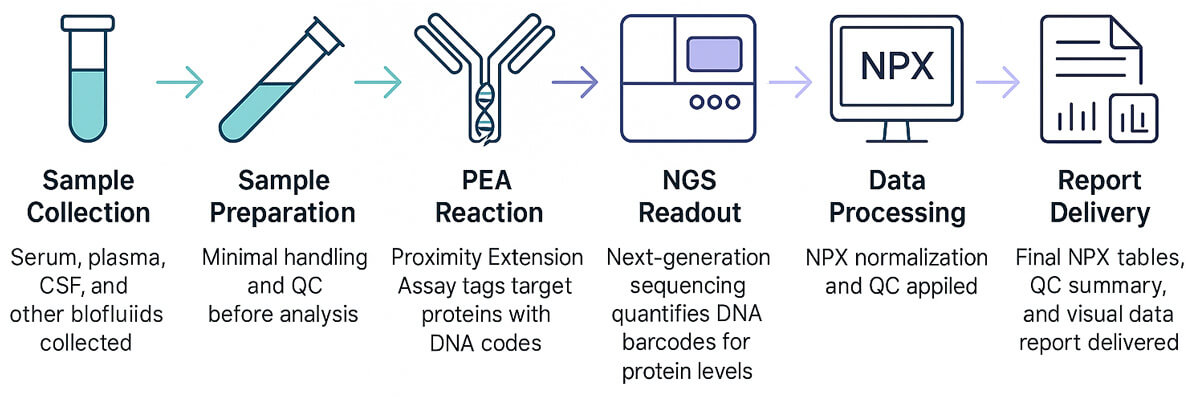
Workflow of Olink Proteomics
Olink Panel Series by Research Focus
We offer disease-focused Olink panel series designed to support a wide range of research areas. Each series targets a specific biological or clinical context, helping scientists choose the most relevant biomarkers for their studies.
Our portfolio covers:
Cardiometabolic health
Explore protein markers linked to diabetes, lipid regulation, and vascular biology.
Cardiovascular disease
Assess cardiac stress, heart failure, and endothelial dysfunction pathways.
Developmental biology
Investigate proteins driving cell growth, differentiation, and tissue formation.
Immune response
Profile cytokines and chemokines to map innate and adaptive immune activation.
Immuno-oncology
Examine checkpoint regulators and tumour–immune system interactions.
Inflammation
Track acute, chronic, and aging-related inflammatory markers.
Detect biomarkers associated with neurodegeneration, synaptic health, and neuroinflammation.
Organ damage
Identify proteins signalling tissue injury or repair in liver, kidney, or other organs.
Sources of Proteins Measured by Olink Proteomics
Olink proteomics panels draw on an exceptionally broad and well-curated library of protein targets, enabling researchers to profile clinically relevant biomarkers with precision. These proteins come from several key sources:
- Human Protein Atlas & Secretome Projects – Core reference datasets ensure panels cover proteins with known biological relevance.
- Tissue- and organ-specific proteins – Includes markers that reflect the activity of distinct organs and systems.
- Inflammation-related proteins – Comprehensive coverage of cytokines, interleukins, and other immune mediators to map immune system dynamics.
- Investigational and validated drug targets – More than 1,000 proteins identified as therapeutic targets, many overlapping with the panels' immune and tissue markers.
- Novel plasma proteins – New biomarkers discovered via complementary proteomics methods and marker-detection platforms.
Applications of Olink Proteomics Technology
Olink proteomics supports a broad spectrum of life science and translational research, giving scientists the tools to move from discovery to application with confidence.
Diagnostic biomarker development
Drives early detection strategies and underpins precision medicine efforts.
Biomarker discovery in drug development
Identifies promising markers during the R&D pipeline to support targeted therapies.
Understanding disease biology
Provides deep proteomic insights to reveal the molecular mechanisms driving disease.
Emerging applications
Expands proteomics into new and rapidly evolving research areas.
Major disease research
Covers seven key disease categories and physiological processes, supporting high-impact studies.
Medical translation
Bridges basic research and clinical practice, helping innovations reach patients faster.
Multi-Omics Integration for Deeper Insights
Our Olink proteomics data is seamlessly integrated with a range of other omics layers, enabling multidimensional insights that go beyond single-omic analysis.
Service combinations include:
Olink + Metagenome / Microbiome
Explore host–microbiome protein–gene interactions in health, disease, and environmental contexts.
Olink + Mass Spectrometry Proteomics
Cross-validate PEA-based and MS-based proteomes to enhance quantitative confidence.
Olink + Single-Cell Transcriptome
Use SPARC-like co-profiling to measure intracellular proteins and mRNA within the same cells.
Correlate non-coding RNA expression with proteomic shifts across pathways.
Identify metabolic changes driven by protein regulation using integrated biomolecule profiling.
Olink + Metagenome + Metabolome
For systems-level mapping of the gut-organ axis, combining microbial gene, metabolite, and host protein data.
These pipelines leverage state-of-the-art statistical, network-based and machine learning strategies to connect DNA, RNA, protein, metabolite, and microbial data into cohesive biological insights.
How Our Olink Proteomics Panel Service Works
- Consultation – Select the right Olink panel or design a custom Olink Flex panel.
- Sample Submission – Send plasma, serum, CSF, urine, or other approved matrices.
- Assay & Analysis – Olink PEA assays run on Signature Q100 or NGS platforms.
- Data Delivery – Get NPX values, heatmaps, volcano plots, and QC reports.
Why Creative Proteomics?
Comprehensive portfolio
Publication-ready outputs
Fast
turnaround
Research-only compliance
The Complete Solution: Olink + Mass Spectrometry
We are not just an Olink provider; we are a world-class mass spectrometry lab. We don't just give you Olink data—we provide the essential next step: gold-standard LC-MS validation, PTM analysis, and unbiased discovery.
Comprehensive Portfolio: Access all Olink platforms (Explore, Target, etc.).
Internal Validation: Seamlessly validate your Olink hits with our in-house mass spec (PRM/SRM) experts.
Go Beyond the Panel: Use our PTM and unbiased proteomics to discover what Olink alone cannot see.
Publication-Ready Data: We deliver a complete, validated, and interpretable dataset, not just an NPX file.
Sample Types Compatible with Olink Proteomics
Olink proteomics technology is designed to work seamlessly with a wide spectrum of sample types, giving researchers flexibility across study designs. Its exceptional sensitivity makes it particularly effective for liquid biopsies and low-abundance protein detection.
Neurological samples
cerebrospinal fluid (CSF) and plaque lysates.
Microparticle fractions
microvesicles and exosomes.
Special formats
dried blood spots and other minimally invasive collection methods.
Sample Requirements
| Sample Type | Volume Required | Notes | Storage |
| Serum | 50 µL | Use clot activator tubes; avoid haemolysis | −80 °C |
| Plasma | 50 µL | EDTA or heparin preferred | −80 °C |
| CSF | 50 µL | Sterile collection | −80 °C |
| Cell Supernatant | 50–100 µL | Clarify by centrifugation | −20–−80 °C |
| Urine (optional) | 100 µL | Midstream, centrifuged | −80 °C |
Sample Submission & Preparation Resources
Before shipping your samples, please review the official submission requirements and preparation guide. Following these recommendations ensures your Olink assay runs smoothly and produces reliable, high-quality data.
Olink Proteomics Sample Submission Guidelines
Comprehensive instructions for preparing, handling, and shipping samples for Olink assays. Learn proper collection, buffer compatibility, and QC requirements to ensure reproducible data.
Olink Sample Preparation Guidelines
Step-by-step guidance for serum, plasma, CSF, and cell lysates. Understand collection methods, storage tips, and key dos & don’ts for reliable proteomic results.
Demo
FAQ – Olink Proteomics Services
1. How does Olink compare to mass spectrometry?
Olink panels complement, rather than replace, mass spectrometry.
Mass spectrometry is ideal for untargeted discovery but can struggle with very low-abundance proteins.
Olink uses PEA technology to deliver greater sensitivity (fg/mL) and multiplexing across hundreds or thousands of pre-validated targets, making it perfect for targeted biomarker studies.
2. Can Olink panels detect rare or low-abundance proteins?
Yes. Olink's PEA platform routinely detects proteins at femtogram per millilitre (fg/mL) levels, capturing markers that many traditional immunoassays and MS approaches cannot reliably quantify.
3. What research stages benefit most from Olink proteomics?
Olink is used from early discovery through translational research:
- Screening broad biomarker sets in discovery studies.
- Validating candidate markers in focused panels.
- Supporting drug development and mechanistic studies with high-quality, reproducible data.
4. Does Olink support cross-study or longitudinal analysis?
Yes. Olink's NPX data format and built-in controls make it straightforward to normalize data across panels, batches, or time points, which is essential for multi-cohort or long-term projects.
5. How customizable are Olink panels?
Our Olink Flex option lets you select 15–30 proteins from 200+ validated assays. This means you can tailor panels to specific hypotheses, disease areas, or exploratory research needs.
6. What makes Olink suitable for liquid biopsy studies?
The platform's ultra-sensitivity and minimal sample input (just 1–6 μL) allow accurate profiling of low-abundance proteins in biofluids like serum, CSF, urine, and saliva—a major advantage for liquid biopsy work.
7. Can Olink data integrate with other omics approaches?
Yes. Olink proteomics data can be combined with transcriptomics, genomics, metabolomics, and microbiome datasets to create rich multi-layer insights for systems biology and biomarker discovery.
8. How does Olink handle automation and reproducibility?
Olink's workflows are automated from sample prep to analysis, minimizing human error and ensuring high reproducibility across runs, labs, and study phases.
9. Are Olink panels validated for animal research?
Yes. The Target 48 Mouse Panel and several other panels are designed or cross-validated for preclinical studies, making Olink valuable for both human and animal models.
10. How does Olink ensure the reliability of its protein measurements?
Each Olink run includes internal controls (incubation, extension, and detection) and external controls to verify assay performance, ensuring the data you receive is publication-ready.
11. What is NPX and why is it important?
NPX (Normalized Protein Expression) is Olink's relative quantification unit. It standardizes data across assays, batches, and instruments, making results easy to compare between studies.
12. Which research fields are adopting Olink most rapidly?
Olink is being widely adopted in immunology, oncology, neurology, and cardiometabolic research. Publications citing Olink-based studies are growing year on year, showing its value in academic, biotech, and pharma pipelines.
13. How does Olink compare to mass spectrometry?
This is the most important question in proteomics today.
- Olink (PEA) is for High-Throughput Screening: It is the best platform for screening thousands of known proteins in very large sample cohorts with minimal sample volume.
- Mass Spectrometry (LC-MS) is for Validation & Discovery: It is the gold standard for validating hits, performing absolute quantification, analyzing PTMs (which Olink cannot do), and discovering novel proteins not on an Olink panel.
The best research combines both. We specialize in an integrated workflow that uses Olink for screening and MS for validation.
References
- Identifying proteomic risk factors for cancer using prospective and exome analyses of 1463 circulating proteins and risk of 19 cancers in the UK Biobank
- Proteomic signatures improve risk prediction for common and rare diseases
- Plasma protein-based organ-specific aging and mortality models unveil diseases as accelerated aging of organismal systems
- Proteomic profiling platforms head to head: Leveraging genetics and clinical traits to compare aptamer- and antibody-based methods