What We Offer: Comprehensive Olink & Multi-Omics Integration
Our services are particularly beneficial for large-scale studies in oncology, immunology, and neurobiology, where understanding complex biological systems is crucial. By integrating multiple omics layers, we help researchers uncover intricate molecular interactions and disease mechanisms, accelerating the development of targeted therapies and personalized medicine.
Core Benefits
Comprehensive Insights: Study protein markers, genomic signals, metabolites, and more in one go, providing a holistic view of biological systems.
Accelerated Discovery: Integrating multiple layers of data improves the reliability of results, enabling faster identification of biomarkers and therapeutic targets.
Cost and Time Efficiency: By leveraging integrated multi-omics approaches, you get more data from a single sample analysis, reducing costs and time associated with separate analyses.
Scalable Solutions: Whether working with small cohorts or large populations, we offer solutions that scale to meet the needs of your research.
Explore Our Multi-Omics Service Offerings
Combining Olink's proteomics with metagenomic sequencing offers insights into how microbial communities influence disease and health outcomes.
Olink & Mass Spectrometry Proteomics:
A hybrid approach where Olink's precise protein profiling is combined with mass spectrometry, providing detailed protein quantification and identification with deep proteomic insights.
Olink & Single-Cell Transcriptome:
This service combines protein expression data with single-cell RNA sequencing, allowing for a deeper understanding of protein-RNA relationships in specific cell populations.
Integrating proteomics with microbiome sequencing to explore the functional relationships between microbial populations and the host proteome.
Analyze the regulation of gene expression through both proteomics and miRNA profiling, shedding light on molecular mechanisms in diseases like cancer and neurological disorders.
Correlate protein data with transcriptomics to map gene expression pathways, essential for understanding cellular responses and disease mechanisms.
A multi-omics approach combining proteomics with metabolomics, enabling a comprehensive understanding of metabolic shifts in various biological processes.
Olink & Metagenome & Metabolome:
A highly integrated approach to understand complex interactions between the microbiome and metabolic pathways, offering insights into health, disease, and drug efficacy.
Olink Proteomics Panel Portfolio – Key Panels
| Panel Name | Quantification | Detection | Proteins | Key Strengths | Typical Applications |
| Olink Explore 3072 | Relative | NGS | 2,926 | Large-scale biomarker discovery across multiple pathways | Inflammation, oncology, neurology, cardiometabolic, organ damage |
| Olink Explore 384 Panels | Relative | NGS | 368–1,472 | Flexible, high-throughput pathway-specific profiling | Same as above |
| Olink Explore 1536 | Relative | NGS | 1,472 | Balanced coverage for broad biomarker screening | Same as above |
| Olink Target 96 | Relative | Signature Q100 | 92 | High-sensitivity biomarker validation | Inflammation, oncology, neurology, cardiometabolic, immune response |
| Olink Target 48 Cytokine Panel | Absolute/Relative | Signature Q100 | 45 | Focused cytokine & immune marker analysis | Cytokine profiling, immune studies |
| Olink Target 48 Mouse Panel | Absolute/Relative | Signature Q100 | 43 | Preclinical cytokine profiling in mouse models | Translational & drug discovery research |
| Olink Flex Custom Panels | Absolute/Relative | Signature Q100 | 21–30 | Custom biomarker selection | Tailored research projects, exploratory studies |
Workflow
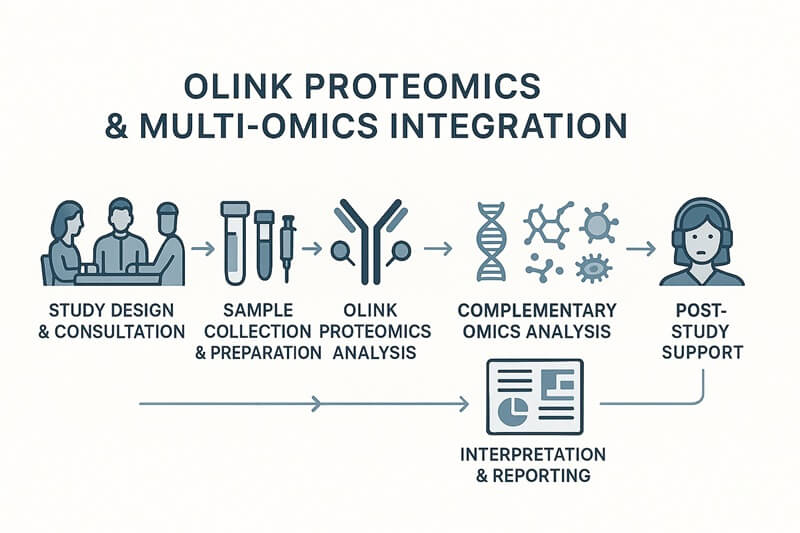
Why Choose Us?
Expertise & Experience:
With years of experience in proteomics and multi-omics technologies, our team helps you design custom experiments that answer your research questions with precision.
Tailored Solutions:
Each service is tailored to your specific research needs. We understand that every project is unique, and our multi-omics integration services are flexible to accommodate a wide range of study designs.
State-of-the-Art Technology:
We use cutting-edge technologies like Olink's Proximity Extension Assay (PEA), next-generation sequencing (NGS), and high-resolution mass spectrometry to ensure high data quality and reproducibility.
Data Integration & Analysis:
Our bioinformatics team ensures that the integrated multi-omics data is processed and analyzed in a way that provides meaningful biological insights, ready for publication.
Sample Requirements
| Sample Type | Volume Required | Notes | Storage |
| Serum | 50 µL | Use clot activator tubes; avoid haemolysis | −80 °C |
| Plasma | 50 µL | EDTA or heparin preferred | −80 °C |
| CSF | 50 µL | Sterile collection | −80 °C |
| Cell Supernatant | 50–100 µL | Clarify by centrifugation | −20–−80 °C |
| Urine (optional) | 100 µL | Midstream, centrifuged | −80 °C |
Delivery
We provide comprehensive deliverables to ensure your research gains maximum value from our Olink & Multi-Omics Integration services:
- Comprehensive Analysis Report – Detailed findings with clear visualizations, pathway analyses, and statistical summaries.
- Raw and Processed Data Files – High-quality datasets for your own downstream analyses.
- Bioinformatics Summary – Interpretation of results, highlighting key biomarkers, pathways, and molecular interactions.
- Customized Data Formats – Delivered in formats compatible with your preferred analysis tools.
- Consultation Session – A post-analysis meeting with our scientists to review findings and answer technical questions.
Frequently Asked Questions (FAQ)
1. How does Olink's Proximity Extension Assay (PEA) support multi-omics integration?
Olink's PEA technology enables high-sensitivity, high-specificity protein quantification using paired antibodies that trigger DNA amplification upon binding to target proteins. This approach allows for the integration of proteomic data with genomic, transcriptomic, and metabolomic datasets, providing a comprehensive view of biological systems.
2. What are the advantages of combining Olink proteomics with other omics technologies?
- Enhanced Biomarker Discovery: Integrating proteomic data with genomic and transcriptomic information facilitates the identification of novel biomarkers and therapeutic targets.
- Holistic Biological Insights: Combining multiple omics layers provides a more comprehensive understanding of disease mechanisms and biological processes.
- Improved Data Reliability: Cross-validation across different omics platforms enhances the robustness and reproducibility of research findings.
3. Which research areas benefit most from multi-omics integration?
Multi-omics integration is particularly valuable in large-scale studies within fields such as oncology, immunology, and neurobiology. It enables researchers to dissect complex biological systems, understand disease mechanisms, and develop personalized therapeutic strategies.
4. What types of samples are suitable for Olink's multi-omics analysis?
Olink's PEA technology is compatible with various sample types, including plasma, serum, tissue lysates, cell culture supernatants, cerebrospinal fluid, and exosomes. This versatility allows for the integration of proteomics data across different biological matrices.
5. How can I get started with Olink's multi-omics services?
To initiate a project or learn more about our services, please contact us to discuss your research needs. Our team will work with you to design a customized multi-omics analysis plan tailored to your specific objectives.
6. How to choose between Olink proteomics and mass spectrometry for biomarker discovery?
Olink proteomics offers high-throughput, high-specificity protein quantification with minimal sample requirements, ideal for large-scale studies and longitudinal biomarker monitoring. Mass spectrometry provides deep proteome coverage and discovery of novel proteins. For targeted, sensitive, and scalable multi-omics analysis services, many researchers combine both technologies for maximum insight.
7. What bioinformatics pipelines are used for multi-omics integration?
We use advanced bioinformatics workflows that integrate proteomics, genomics, transcriptomics, metabolomics, and microbiome data. This includes normalization, differential expression analysis, pathway enrichment, network analysis, and machine learning models for biomarker prediction.
8. What is the typical turnaround time for an Olink multi-omics CRO project?
Turnaround time depends on factors such as project scope, number of omics layers, sample size, and data complexity. Standard Olink proteomics projects are generally completed faster than fully integrated multi-omics studies, but exact timelines are confirmed during project planning to ensure they align with your research needs.
9. How much sample is required for Olink proteomics and multi-omics analysis?
Olink PEA technology requires as little as 1 µL of plasma or serum, making it highly efficient for precious samples. Other omics layers, such as metabolomics and metagenomics, have specific requirements, which we provide upon project planning.
10. Which research fields benefit most from multi-omics integration?
Multi-omics CRO solutions are particularly impactful in oncology, immunology, neurology, infectious diseases, and metabolic disorders, enabling researchers to uncover complex molecular interactions and disease mechanisms.
11. Can I integrate existing datasets with new Olink proteomics results?
Yes, our bioinformatics team can integrate your historical genomics, transcriptomics, or metabolomics datasets with newly generated Olink proteomics data to build a comprehensive, multi-layer biological profile.
Case Study: Integrating Olink Proteomics with Multi-Omics Analysis in Cardiovascular Disease Research
Eldjarn GH, et al,. Large-scale plasma proteomics comparisons through genetics and disease associations. Nature. 2023
- Study Overview
- Multi-Omics Integration
- Implications for Research and Clinical Applications
- Conclusion
A recent large-scale study published in Nature Communications utilized Olink's high-sensitivity proteomics platform to investigate plasma protein biomarkers associated with heart failure (HF) development. The study measured 4,877 plasma proteins in 13,900 HF-free individuals across diverse age groups and geographies. The researchers identified 37 proteins consistently associated with incident HF, independent of traditional risk factors. Additionally, Mendelian randomization analyses suggested causal effects of 10 proteins on HF, HF risk factors, or left ventricular size and function, including matricellular proteins such as SPON1 and MFAP4 .
In this study, the integration of proteomics data with genetic information was a key strength. The researchers performed genome-wide association studies (GWAS) to identify protein quantitative trait loci (pQTLs) and examined their associations with HF. This multi-omics approach allowed for a deeper understanding of the genetic underpinnings of HF and the identification of novel biomarkers and therapeutic targets.
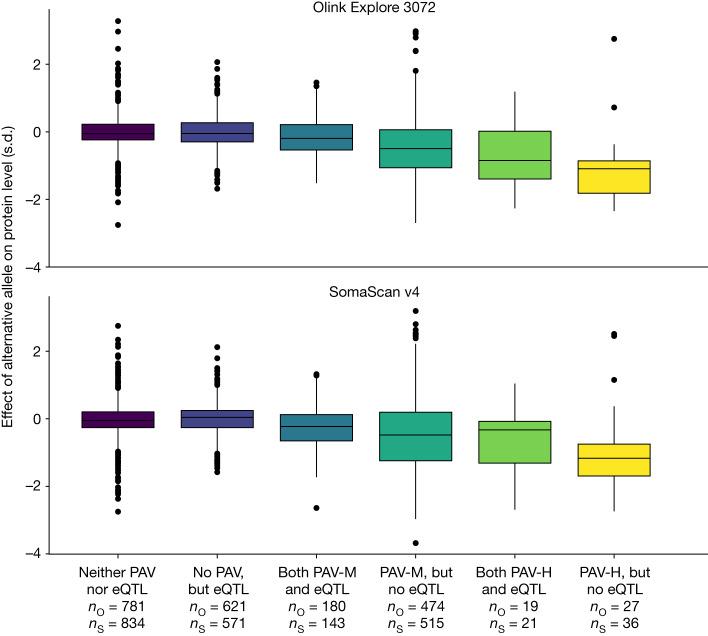 Comparison of alternative-allele detection by Olink Explore 3072 (top) vs SomaScan v4 (bottom). Each box represents the effect size (log₂ fold change) for assays detecting alternative alleles across multiple proteins and genotypes.
Comparison of alternative-allele detection by Olink Explore 3072 (top) vs SomaScan v4 (bottom). Each box represents the effect size (log₂ fold change) for assays detecting alternative alleles across multiple proteins and genotypes.
The findings from this study highlight the potential of combining Olink's proteomics platform with other omics technologies to uncover complex biological mechanisms. By integrating proteomic data with genetic information, researchers can identify biomarkers that are not only associated with disease but also have causal relationships. This approach can inform drug discovery and development, as well as contribute to personalized medicine strategies.
This case study demonstrates the power of multi-omics integration in advancing our understanding of cardiovascular diseases. The use of Olink's proteomics platform, combined with genetic analyses, provides a comprehensive view of the molecular factors contributing to disease development. Such integrative approaches are essential for identifying novel biomarkers and therapeutic targets, ultimately leading to improved patient outcomes.
References
- Chang R, Wang H, et al. Olink proteomics reveals TNFRSF9 as a biomarker for abdominal aortic aneurysms. iScience. 2025 Jun 5;28(7):112828.
- Ekaterina E. Esenkova, et al. Unbiased multi-omics network-based data integration allows clinically relevant outcome-predicting clustering of individuals with heart failure. medRxiv 2025.01.28.25321241;
- Xu, Y., Ritchie, S.C., Liang, Y. et al. An atlas of genetic scores to predict multi-omic traits. Nature 616, 123–131 (2023).
- Wu, TS., Hsiao, TH., Chen, CH. et al. ARTN and CCL23 predicted chemosensitivity in acute myeloid leukemia: an Olink® proteomics approach. Clin Proteom 22, 3 (2025).
- Raih, M., Sage, E., Ali, Q.,et al. (2025). The impact of mutations on TP53 protein and MicroRNA expression in HNSCC: Novel insights for diagnostic and therapeutic strategies. PLoS One, 20(5), e0307859.
- Sharew, N., Arsano, Y., Ahmed, et al. (2025). Multi-omics approaches for understanding gene-environment interactions in noncommunicable diseases: Techniques, translation, and equity issues. Human Genomics, 19(), 1-27.