- Overview
- Panels List
- Applications
- Workflow
- Why Creative Proteomics
- Demo
- Sample Requirements
- Case Study
- FAQ
- Sample Submission Pack
Why Use Olink Proteomics Panels for Drug Development?
Olink proteomics offers a unique combination of precision, scalability, and integration capabilities that address key bottlenecks in drug development—from target discovery to clinical biomarker validation. Its ability to deliver high-quality, multidimensional protein data in challenging matrices makes it an indispensable tool for modern therapeutic development. Here are the key reasons why Olink proteomics technology is highly valuable for drug development:
- High Sensitivity: Detect sensitivity reaching fg/mL levels, capable of measuring low-abundance pharmacodynamic biomarkers. This enables monitoring of subtle drug effects and low-abundance signaling proteins critical for understanding mechanism of action.
- High Multiplexing: Simultaneous quantification of 48-3,072 proteins from a single minimal sample volume. This provides comprehensive signaling pathway coverage for unbiased biomarker discovery and polypharmacology assessment.
- Minimal Sample Requirement: Only 1-6 μL of plasma/serum needed for full proteomic profiling. This preserves precious clinical trial samples for additional analyses and enables pediatric/rare disease studies.
- Excellent Data Quality: Low intra-assay (<10%) and inter-assay (<20%) coefficients of variation. This generates reliable, reproducible data meeting regulatory standards for biomarker qualification.
- Wide Dynamic Range: 10-log dynamic range covering fg/mL to mg/mL protein concentrations. This captures both high-abundance clinical chemistry markers and low-abundance signaling molecules without dilution.
Selecting the Right Panel for Protein Drug Development
The most appropriate panels are those explicitly designed to measure predefined sets of proteins relevant to specific biological processes, disease areas, or pathways, allowing for focused hypothesis testing. Panels were selected based on their:
- Specificity: They target proteins known to be biomarkers in well-defined biological areas (e.g., inflammation, cardiometabolic health, oncology).
- Multiplexing Capability: They allow for the simultaneous measurement of multiple proteins from a single, small sample volume, which is essential for comprehensive profiling.
- Relevance to Disease Mechanisms: The panels are constructed around key pathways and processes that are frequently dysregulated in disease, making them directly applicable to biomarker discovery and validation.
Here is a list of recommended panels we offer:
Table. List of Olink Panels
Additional Options
In addition to the panels above, we also offer the following panels:
- Target 96
- Custom Olink Flex systems
Our team can assist in designing an integrated Olink–Drug Development strategy tailored to your project goals.
Applications of Drug Development
- Target Discovery and Validation
Olink facilitates novel drug target identification through protein quantitative trait locus (pQTL) studies that connect genetic variants to protein expression and disease phenotypes. By analyzing large population cohorts, researchers can identify causal protein-disease relationships with higher confidence than genomic approaches alone, significantly improving target validation success rates.
- Preclinical Development
In preclinical studies, Olink panels enable comprehensive mechanism of action (MOA) studies, safety assessment, and pharmacokinetic/pharmacodynamic (PK/PD) modeling. The ability to profile thousands of proteins simultaneously helps identify both intended drug effects and unexpected biological consequences early in development, reducing late-stage failures.
- Dedicated Analytical Assays
The Olink Flex platform enables the creation of customized panels targeting 15-21 proteins specifically aligned with a compound's mechanism of action. These tailored panels can be optimized and validated as dedicated analytical assays for research cohort stratification and longitudinal biomarker response assessment, leveraging the same underlying PEA technology that facilitated the initial biomarker discovery.
Workflow of Drug Development
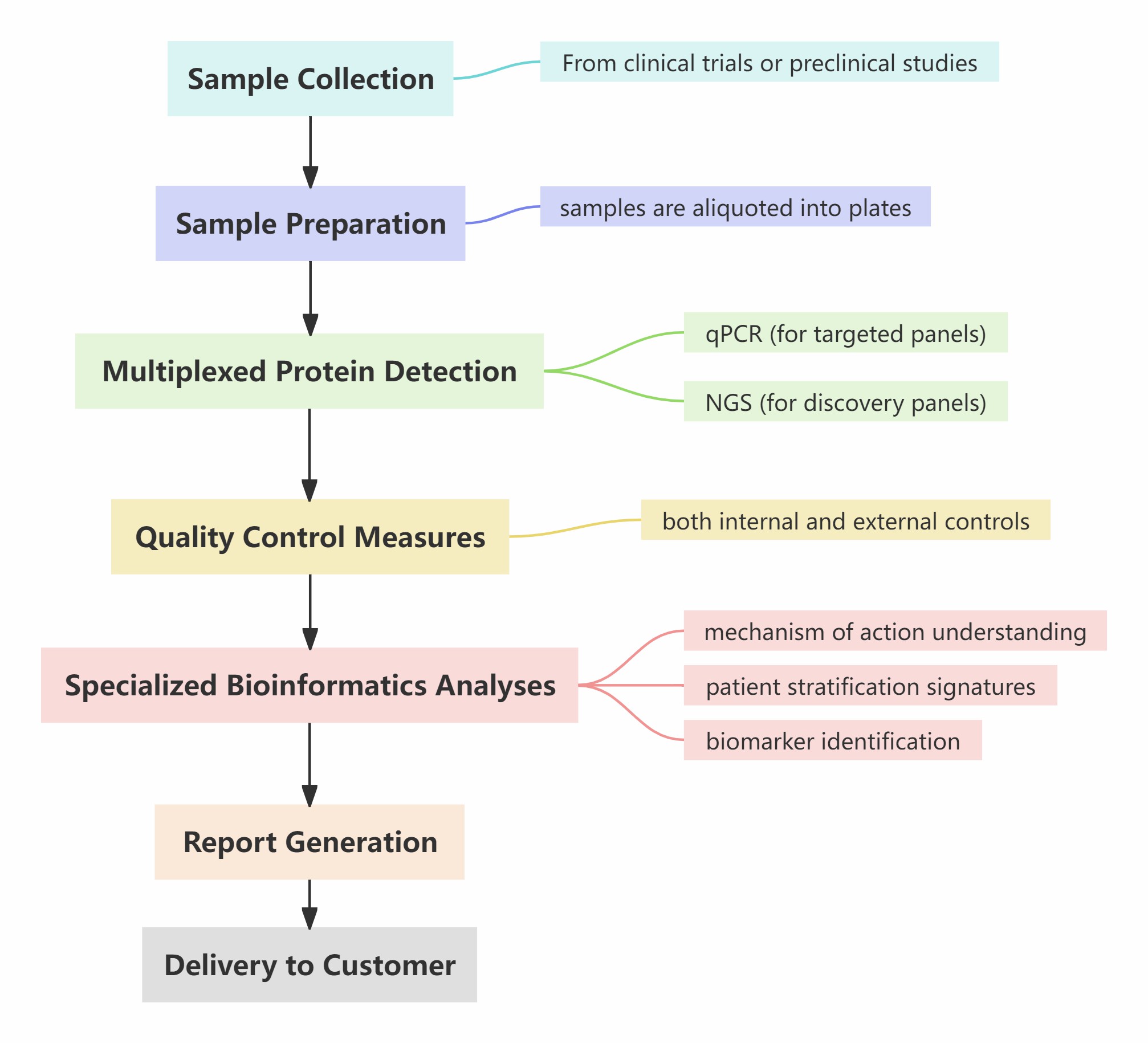 Figure 1: Workflow of Olink' s Drug Development Service.
Figure 1: Workflow of Olink' s Drug Development Service.
Why Creativ Proteomics
High-Plex, High-Sensitivity Target Discovery
This high-plex, ultra-sensitive approach (detection down to fg/mL levels) is crucial for uncovering novel drug targets, such as through protein quantitative trait loci (pQTL) studies, and for understanding complex biology by covering 100% of major signaling pathways.
Accelerated Mechanistic Understanding and Safety Profiling
We provide deep insights into a compound's mechanism of action (MoA), biocompatibility (Safety), pharmacokinetics (PK), pharmacodynamics (PD), and optimal dosage (Dose). By revealing the protein-level changes induced by an intervention, it helps explain variability in compound response and observed biological effects among subjects, facilitating stratified research approaches and de-risking development.
Flexibility for Hypothesis Testing
Our service allows researchers to create custom panels by selecting 15-21 proteins from a library of nearly 200 low-abundance markers. This offers a cost-effective and flexible solution for targeted verification of specific hypotheses in drug development without the need for large-scale discovery experiments, using only 1 µl of sample.
Demo Results: Olink-Drug Development
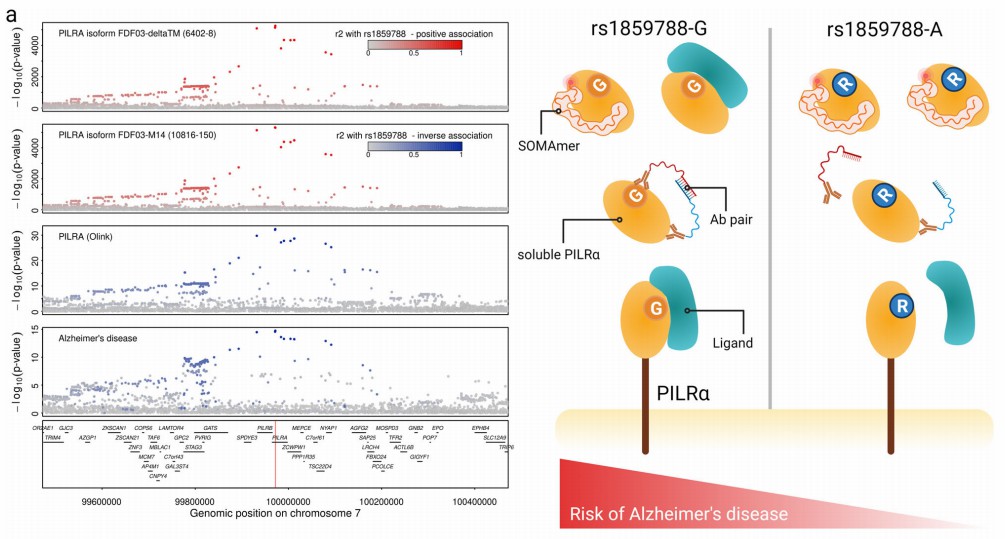 Figure 2: Regional association plots for paired immunoglobulin-like type 2 receptor alpha (PILRα) measured by SomaScan (top rows) and Olink. (Pietzner, M., et al. 2021)
Figure 2: Regional association plots for paired immunoglobulin-like type 2 receptor alpha (PILRα) measured by SomaScan (top rows) and Olink. (Pietzner, M., et al. 2021)
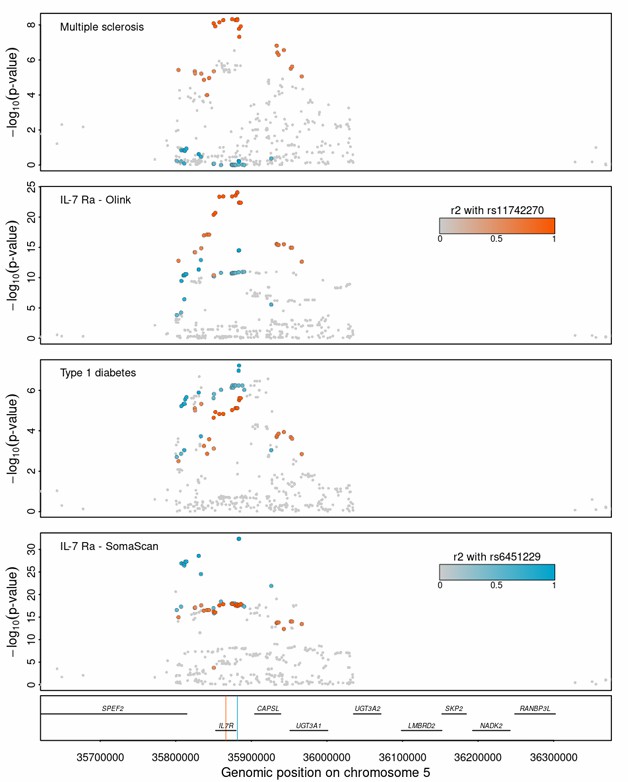 Figure 3: Regional association plots for multiple sclerosis, type 1 diabetes, and Interleukin 7 receptor subunit alpha (IL-7 Ra) measured by Olink and SomaScan. (Kuliesius, J., et al. 2025)
Figure 3: Regional association plots for multiple sclerosis, type 1 diabetes, and Interleukin 7 receptor subunit alpha (IL-7 Ra) measured by Olink and SomaScan. (Kuliesius, J., et al. 2025)
Sample Requirements for Drug Development
1. Sample Types
- The platform is optimized for blood-derived samples including plasma (various anticoagulants) and serum, which are most commonly collected in clinical trials. Other biofluids such as CSF, urine, and tissue culture supernatants are also compatible.
2. Volume Requirements
- Typical requirements range from 1 μL for targeted panels to 6 μL for comprehensive 3,000-protein profiling.
3. Sample Storage and Shipping
- Standard protocols for clinical sample processing should be followed, with storage at -80°C and shipping on dry ice to maintain protein integrity.
Case Study

Title: Prognostic tools and candidate drugs based on plasma proteomics of patients with severe COVID-19 complications
Journal: Nat Commun
Year: 2022
- Background
- Methods
- Results
The COVID-19 pandemic has placed an immense burden on healthcare systems worldwide, highlighting the urgent need for early prognostic tools to identify patients at risk of severe complications. This study aimed to address this gap by profiling plasma proteins from 50 severe and 50 mild-moderate COVID-19 patients, alongside 50 healthy controls. The researchers identified 375 differentially expressed proteins (DEPs) in severe cases, implicating these proteins in the pathogenesis of COVID-19 and suggesting their potential as targets for therapeutic interventions. Additionally, the study proposed a 12-plasma protein signature and a model incorporating seven routine clinical tests to predict disease severity and survival, validated in an independent cohort. These findings offer a foundation for personalized management of SARS-CoV-2 infections.
The study utilized Olink's proximity extension assay (PEA) technology to profile 893 plasma proteins across ten targeted panels, each focusing on specific biological processes or disease pathways. Plasma samples from patients and controls were analyzed using these panels, and quality control was performed using Olink's NPX Manager software. Proteins with expression values reported as log2 Normalized Protein Expression (NPX) were included in the analysis. Differential expression analysis was conducted using the limma package in R, accounting for covariates such as obesity, sex, age, ethnicity, heart rate, and oxygen saturation (SpO₂). Proteins with a fold change >1.25 and a false discovery rate (FDR) <0.1 were considered differentially expressed. Unsupervised hierarchical clustering and principal component analysis (PCA) were employed to visualize group separations based on protein expression patterns.
Olink's multi-panel proteomic analysis revealed significant dysregulation of plasma proteins in severe COVID-19 patients. Specifically, 375 DEPs were identified in severe versus mild-moderate cases, with the majority being upregulated. Unsupervised clustering of the combined protein data (893 unique proteins) clearly distinguished severe cases from mild-moderate and control groups. Functional annotation categorized these DEPs into 11 groups, including cytokines, immune evasion markers, coagulopathy-related proteins, and indicators of organ damage. Notably, C-reactive protein (CRP) exhibited the strongest correlation with DEPs (82%), followed by creatinine (64%) and urea (56%). The 12-protein signature derived from this data demonstrated exceptional accuracy in predicting disease severity, with 100% specificity and 98% sensitivity (AUC: 0.999). This signature was validated in an independent cohort, where it effectively predicted severity and mortality outcomes up to 28 days post-admission. These results underscore the utility of Olink-based proteomic profiling in identifying robust biomarkers for COVID-19 prognosis.
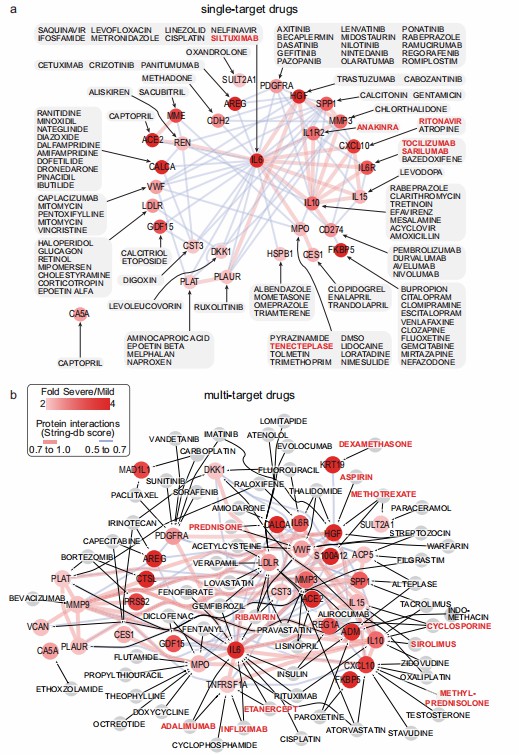 Figure 4: Drug–protein interactions of upregulated plasma proteins in severe COVID-19 patients. (Al-Nesf, M.A.Y., et al. 2022)
Figure 4: Drug–protein interactions of upregulated plasma proteins in severe COVID-19 patients. (Al-Nesf, M.A.Y., et al. 2022)
FAQs
How does Olink support multi-omics and real-world data integration?
Olink data can be integrated with genomic and clinical datasets to uncover deeper biological insights. For example, the UK Biobank's pharmaceutical proteomics project uses Olink to generate large-scale protein-genomic data from 56,000 samples, enabling global research into disease mechanisms and drug targets.
What sample types are compatible with Olink assays?
Olink assays work well with a variety of sample types, including:
- Plasma and serum (especially advantageous for low-abundance proteins).
- Other body fluids (e.g., CSF, urine).
- Tissue extracts (with appropriate validation).
References
- Pietzner, M., Wheeler, E., Carrasco-Zanini, J. et al. Synergistic insights into human health from aptamer- and antibody-based proteomic profiling. Nat Commun 12, 6822 (2021).
- Kuliesius, J., Timmers, P.R.H.J., Navarro, P. et al. Efficient candidate drug target discovery through proteogenomics in a Scottish cohort. Commun Biol 8, 1300 (2025).
- Al-Nesf, M.A.Y., Abdesselem, H.B., Bensmail, I. et al. Prognostic tools and candidate drugs based on plasma proteomics of patients with severe COVID-19 complications. Nat Commun 13, 946 (2022).