- Overview
- Panels List
- Applications
- Workflow
- Why Creative Proteomics
- Demo
- Sample Requirements
- FAQ
- Sample Submission Pack
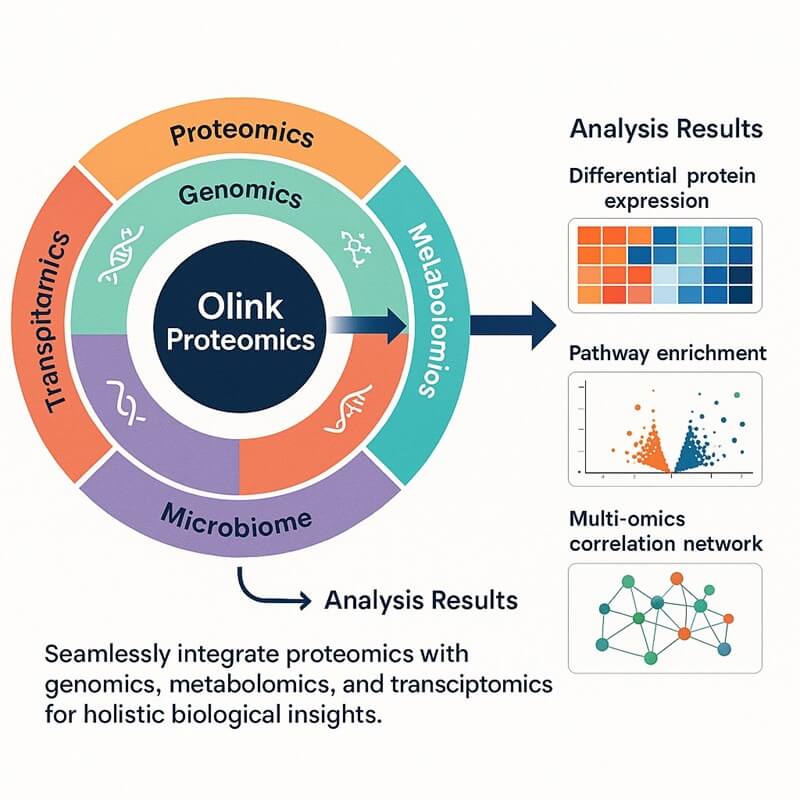
Why Integrate Olink Proteomics with Metagenome and Metabolome?
The integration of Olink and transcriptome provides a transformative multi-omics framework for deciphering complex biological systems. This integrated service addresses limitations of single-omics approaches by capturing functional interactions across host molecular pathways, microbiome activities, and metabolic states. Single-omics studies reveal potential biological states, but the Olink-metabolome-Metagenome triad quantifies active physiological processes.
Olink Technology
Proximity Extension Assay (PEA)
1) Antibody pairs conjugate to DNA oligonucleotides.
2) Antibody pairs bind to target protein.
3) Upon co-localization, the DNA strands hybridize and extend into amplifiable templates.
4) Quantifiable DNA signals via qPCR or NGS.
Metagenome sequencing
1) Microbial sample collection.
2) DNA extraction.
3) Sequencing.
4) De novo assembly or align to reference genome.
5) Metagenome amplicon sequencing or metagenomics random shortgun sequencing.
Metabolome
1) Metabolite Detection and Quantification:
- Mass Spectrometry (MS) coupled with chromatography.
- Nuclear Magnetic Resonance (NMR)
2) Data Processing and Metabolite Identification.
3) Analytical Strategies: Untargeted metabolomics, targeted metabolomics and widely targeted metabolomics.
4) Biological Interpretation: Pathway analysis and multi-omics Integration.
Advantages
- Minimal Sample Requirements: Requires only 1–6 µL of sample (e.g., serum, plasma, CSF) for simultaneous quantification of 1,500–3,000 proteins.
- Unprecedented Sensitivity and Specificity: Detects low-abundance proteins at fg/mL levels in complex biofluids, overcoming limitations of mass spectrometry. Dual antibody-DNA recognition minimizes cross-reactivity and false positives.
- Cross-omics validation: Olink proteins and metabolome metabolites co-validate pathways.
- Holistic insights: Links host proteome, metabolome, and microbiome in diseases.
Olink & Metagenome & Metabolome Platform: Selecting the Right Panel
Recommended Panels for Integrated Analysis
We recommend prioritizing the Target 96 Metabolism, Immuno-Response, Inflammatory, and Organ Damage panels, complemented by targeted Custom Flex panels like the UKB T2D Panel or Inflammation in Aging Panel when disease-specific. Because these options provide focused, high-quality data on proteins intimately involved in metabolic regulation, immune response modulation, inflammation, and tissue homeostasis.
Here is a list of recommended panels we offer:
Table. List of Olink Panels
| Platforms | Recommended Olink Panels |
| Explore 384 | Olink Explore 384 Inflammation |
| Olink Explore 384 / Inflammation Ⅱ | |
| Target 96 | Olink Target 96 Immuno-Oncology |
| Olink Target 96 Metabolism | |
| Olink Target 96 Inflammatory | |
| Olink Target 96 Organ Damage Panel | |
| Custom Olink Flex systems | Inflammation in aging Panel |
| UKB type 2 diabetes Panel | |
| Cytokine storm Panel |
Additional Options
In addition to the panels above, we also offer:
- Explore HT
- Explore 3072 (3072 proteins)
- Explore 1536 (1536 proteins)
- Explore 384
- Target 96
- Olink Target 96 Cell Regulation Panel
- Olink Target 96 Oncology II Panel
- Olink Target 96 Oncology III Panel
- Olink Target 96 Cardiovascular II Panel
- Olink Target 96 Cardiovascular III Panel
- Olink Target 96 Neurology
- Olink Target 96 Neuro Exploratory
- Olink Target 96 Cardiometabolic
- Olink Target 96 Immune Response
- Olink Target 96 Mouse Exploratory Panel
- Olink Target 96 Development Panel
- Target 48
- Custom Olink Flex systems
Our team can assist in designing an integrated Olink–Metagenome-Metabolome strategy tailored to your project goals.
Applications
These applications leverage multi-omics synergy to map interactions across biological scales, advancing mechanistic research in precision health and disease biology:
Host-Microbiome Interaction Networks
- This approach deciphers how gut microbial communities (metagenome) influence host physiology by producing metabolites that modulate protein expression (proteomics).
Multi-Omic Drivers of Complex Traits
- Integrated analysis identifies how genetic factors and microbial features jointly explain individual variations in circulating protein levels. Genetic variants primarily regulate immune-related proteins, while microbes dominate metabolic protein profiles.
Functional Biomarker Discovery
- Combining proteomics with metagenome/metabolome uncovers early-prediction signatures for diseases like Parkinson' s or cancer.
Causal Target Prioritization
- Mendelian randomization integrates genomic data with proteomics to identify proteins causally linked to diseases. Metabolome and metagenome data then validate whether microbial or metabolic pathways regulate these targets, accelerating therapeutic development.
Intervention Response Profiling
- Monitoring protein, metabolite, and microbial dynamics during dietary or therapeutic interventions quantifies biological impact.
Workflow of Olink & Metagenome & Metabolome
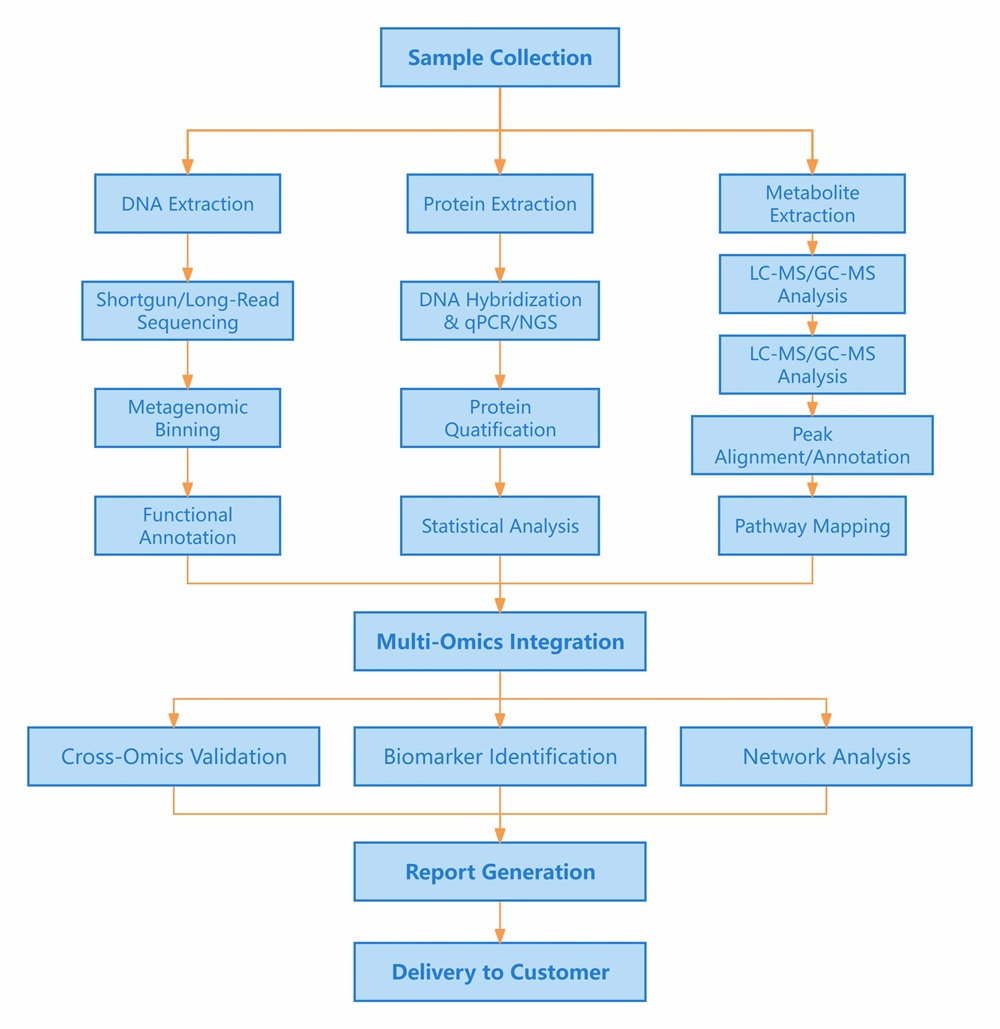 Figure 1: Workflow of integrated Olink proteomics, Metagenome and Metabolome.
Figure 1: Workflow of integrated Olink proteomics, Metagenome and Metabolome.
Why Creativ Proteomics
Advanced Multi-Omics Integration Capabilities
- Cross-Platform Data Harmonization: Ability to integrate data from Olink (targeted proteomics), metabolomics, and proteomics into a unified analysis pipeline. This includes reconciling technical differences.
- Joint Pathway and Network Analysis: Expertise in identifying multi-layer molecular interactions (e.g., protein-metabolite correlations in disease mechanisms).
Cutting-Edge Technology and Platforms
- High-Sensitivity Platforms: Proficiency in Explore HT (5,300+ proteins, 2μl sample volume) or Target 48/96 for focused studies. Use of high-resolution mass spectrometry for broad metabolite coverage. Standard workflows for deep proteome profiling.
Robust Bioinformatics
- Integrated Knowledge Bases: Use of biomedical knowledge graphs to annotate proteins/metabolites and prioritize biomarkers.
- Data Reproducibility
End-to-End Quality Control & Standardization
- Cross-Platform QC: NPX normalization and sample/plate controls for Olink.
- Orthogonal Validation: Use of multiple platforms to verify key biomarkers.
Domain-Specific Expertise & Customization
- Disease-Focused Panels: Customized multi-omics panels for diseases like cancer, cardiovascular disorders, or neurodegeneration (e.g., combining Olink Inflammation Panel with metabolomics for stroke biomarker discovery).
- Sample Type Versatility: Experience with diverse samples (serum, plasma, CSF, tissue) and low-volume handling (critical for pediatric or longitudinal studies)
Demo Results
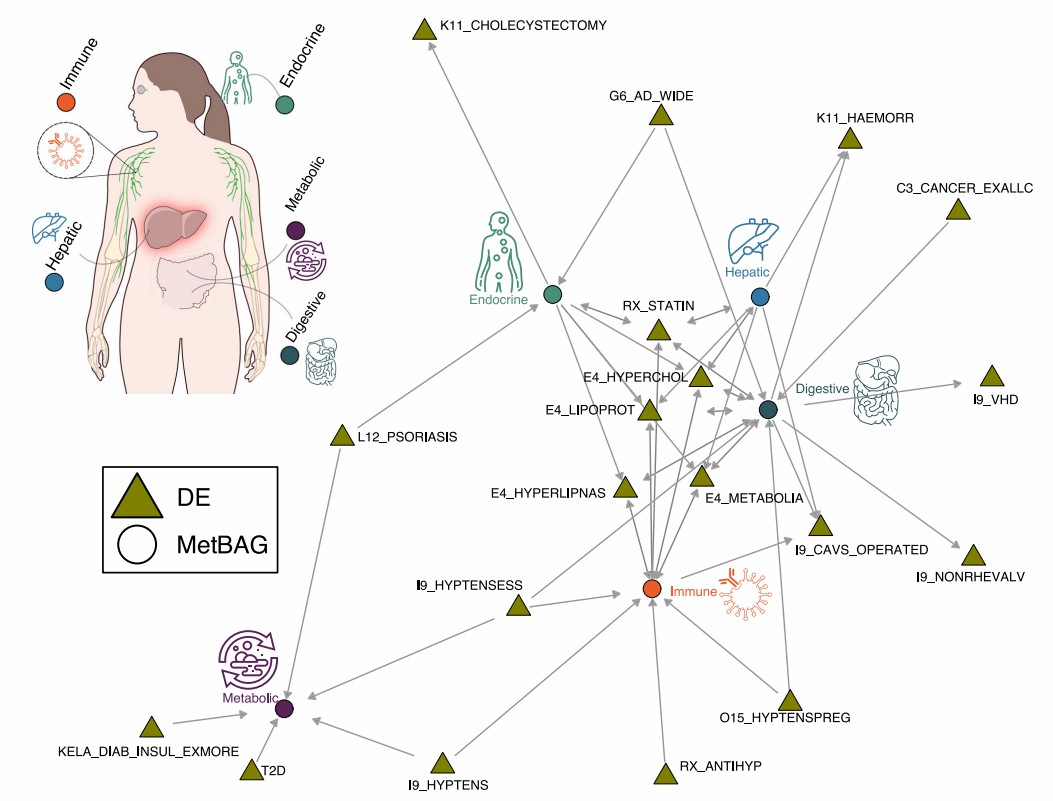 Figure 2: Causal networks between the 5 MetBAGs and 525 Des. (The MULTI consortium., et al. 2025)
Figure 2: Causal networks between the 5 MetBAGs and 525 Des. (The MULTI consortium., et al. 2025)
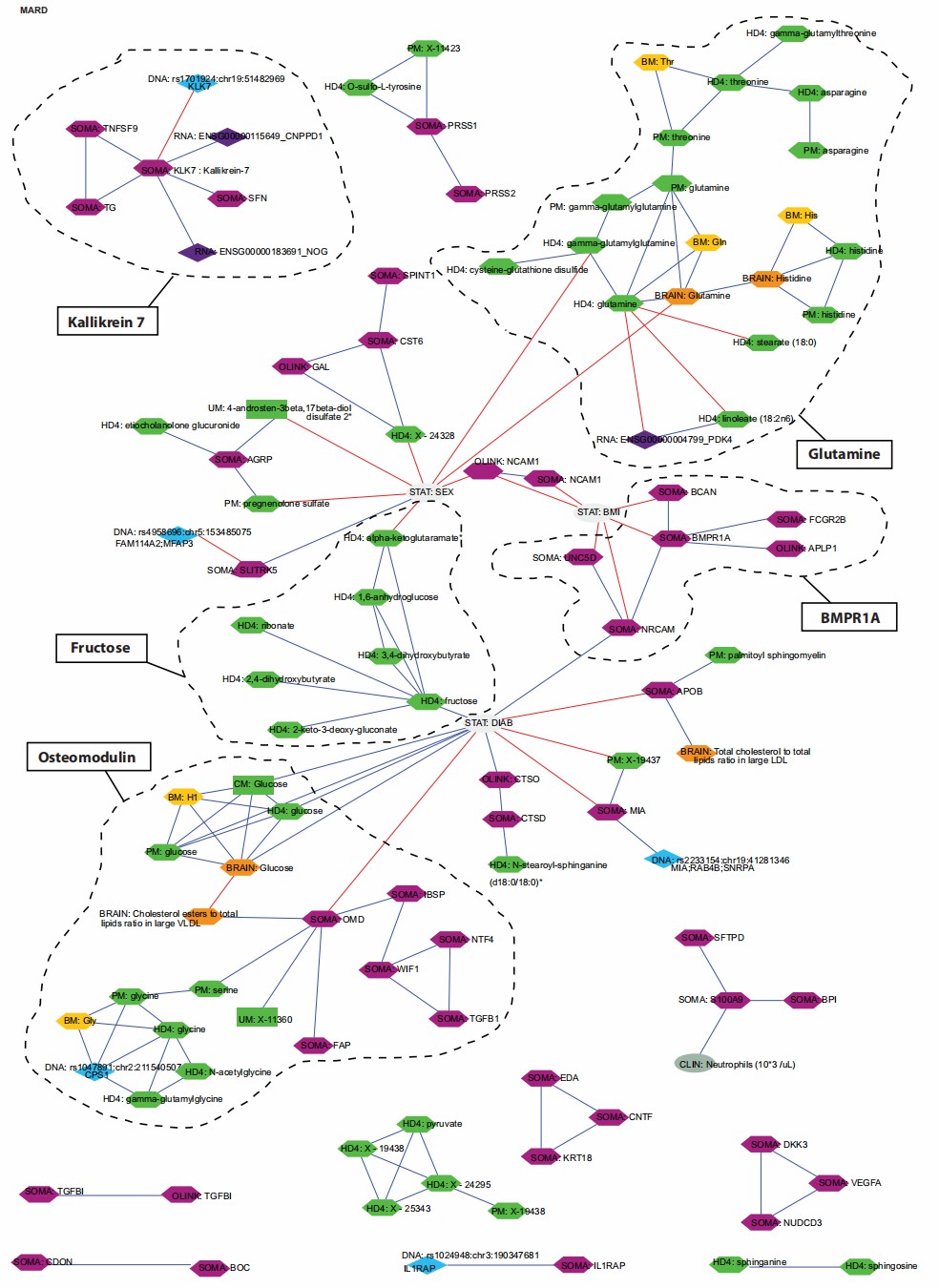 Figure 3: Multiomics interaction network of protein and metabolite signatures in MARD-associated clusters. (Halama, A., et al. 2024)
Figure 3: Multiomics interaction network of protein and metabolite signatures in MARD-associated clusters. (Halama, A., et al. 2024)
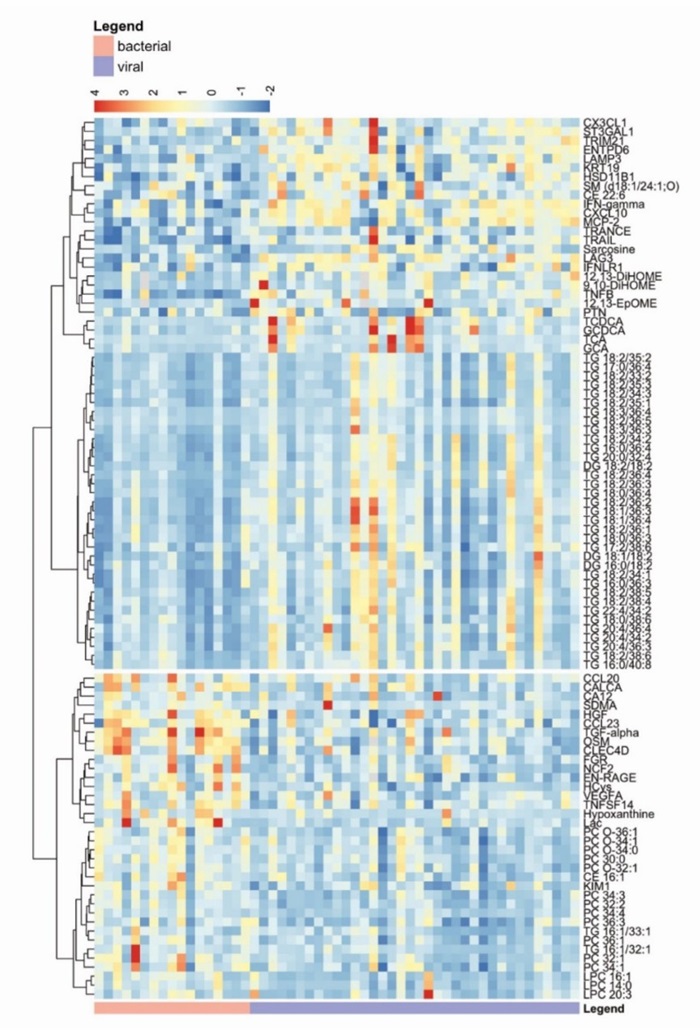 Figure 4: Co-expression of significantly altered lipids, metabolites and proteinss. (Rischke, S., et al. 2025)
Figure 4: Co-expression of significantly altered lipids, metabolites and proteinss. (Rischke, S., et al. 2025)
Sample Requirements
| Step | Olink Proteomics | Metagenomics (Shotgun Sequencing) | Metabolomics |
| Sample Types | Serum, plasma, CSF, urine, tissue lysates, cell supernatants | Body fluids (BALF, CSF, blood), tissue, sterile swabs | Serum, plasma, urine, CSF |
| Volume |
|
|
≥50–100 μL (serum/plasma); ≥200 μL (urine) |
| Avoid | Hemolysis, repeated freeze-thaw, or samples exposed to fluorescein (interferes with qPCR) | Hemolysis (increases human DNA background), delays in processing, or non-sterile collection | Additives (e.g., sodium azide), hemolysis, or room-temperature exposure >30min |
| Storage Temperature | –80°C (no dry ice sublimation) | –80°C (vapor-phase LN₂ preferred) | –80°C (avoid frost-free freezers) |
| Transport | Ship on dry ice | ||
Notes
- Cross-Contamination: Use aerosol-resistant tips during aliquoting.
- Inconsistent Collection: Use identical anticoagulants (e.g., EDTA for all plasma).
- Thawing for Sub-aliquoting: Pre-plan aliquot sizes to avoid thawing main stock.
FAQs
What sample types are compatible across these technologies?
The Olink platform is validated for EDTA plasma and serum, with broad compatibility extending to CSF, saliva, tissue lysates, and exosomal extracts. Metabolomics supports diverse sample types including serum/plasma, cells, tissues, and microbial cultures. In contrast, metagenomics specifically requires microbial genomic DNA derived from stool, environmental samples, or biopsy tissues to reconstruct genome-scale metabolic models for predicting microbial community interactions. For integrated multi-omics workflows, sample fractionation is critical. Biofluids can be aliquoted for metabolomics (prioritizing immediate metabolite extraction), Olink proteomics (stable proteins), and metagenomics (DNA from microbial-enriched fractions), ensuring analyte integrity across assays.
How to minimize batch effects in multi-omics studies?
To minimize batch effects in multi-omics studies, a three-pronged approach is essential:
- Reference sample-based normalization: For Olink and metagenome, use 8–16 bridging reference samples across batches to convert absolute feature values into ratios relative to the reference. This can effectively decouple batch-specific technical variations from biological signals.
- Quality control (QC) integration in metabolomics: Include pooled QC samples processed alongside study samples in each batch. These QCs enable normalization via probabilistic quotient normalization or similar methods, correcting for instrumental drift and matrix effects.
- Automated workflows: Adopt standardized, high-throughput platforms such as EMR-lipid 96-well plates for metabolomics to ensure consistent sample handling, extraction, and analysis. Automation reduces human-induced variability and enhances reproducibility across batches.
What is the optimal order of sample processing?
The optimal order for processing multi-omics samples prioritizes metabolomics first due to the extreme lability of metabolites, requiring immediate quenching to halt enzymatic activity and prevent degradation. Olink proteomics follows, as proteins remain relatively stable during short-term storage at –80°C. Microbial genomics is processed last, since microbial DNA retains integrity in preservative buffers for weeks, allowing flexible storage at 4°C or room temperature with stabilization agents.
This sequential workflow (metabolites → proteins → microbial DNA) minimizes analyte degradation, leverages differential stability across biomolecule classes, and ensures data fidelity for integrated multi-omics analysis.
How to integrate heterogeneous data types?
Effective integration of heterogeneous data types requires platform-specific normalization followed by advanced cross-omics modeling.
For normalization:
- Olink proteomics employs NPX on a log₂ scale to minimize plate- and run-specific biases while preserving relative quantification.
- Metabolomics leverages PQN to correct for sample dilution variations or uses internal standards to calibrate absolute concentrations against technical noise.
- Metagenomics normalizes microbial gene counts via TPM to account for sequencing depth and gene length biases, enabling accurate comparisons across microbiome samples.
References
- The MULTI consortium., Anagnostakis, F., Ko, S. et al. Multi-organ metabolome biological age implicates cardiometabolic conditions and mortality risk. Nat Commun 16, 4871 (2025).
- Halama, A., Zaghlool, S., Thareja, G. et al. A roadmap to the molecular human linking multiomics with population traits and diabetes subtypes. Nat Commun 15, 7111 (2024).
- Rischke, S., Gurke, R., Zielbauer, AS. et al. Proteomic, metabolomic and lipidomic profiles in community acquired pneumonia for differentiating viral and bacterial infections. Sci Rep 15, 1922 (2025).