- Overview
- Panels List
- Applications
- Workflow
- Why Creative Proteomics
- Demo
- Sample Requirements
- Case Study
- FAQ
- Sample Submission Pack
Why Discover Protein Biomarker with Olink Panels?
Olink panels excel in Protein Biomarker Profiling by offering unparalleled scalability (>5,400 proteins simultaneously), minimal sample requirements (ideal for longitudinal/biobank studies), and rigor sufficient for clinical validation. Its PEA technology avoids the limitations of mass spectrometry (e.g., loss of low-abundance proteins) and ELISA (low throughput). By delivering reproducible, high-quality data, Olink enables an efficient transition from biomarker discovery to the development of practical analytical tools.
Selecting the Right Panel for Protein Biomarker Profiling
For robust and interpretable protein biomarker profiling, the Target 96 series represents the most balanced and recommended option. The selection of the panels is based on the following criteria:
- Breadth of Covered Biomarkers
The Target 96 panels offer broad disease area coverage. This makes them ideal for general profiling purposes where the goal is to identify significant protein changes across multiple biological pathways without being overly restricted to a single, narrow focus.
- Established Disease Associations
Most of the Target 96 panels have established disease associations. This is a key advantage for biomarker profiling, as it means the measured proteins have known relevance to human diseases, facilitating the interpretation of results and linking findings to existing biological knowledge.
- Functional Focus
Unlike the very high-plex Explore HT panels (1536/3072), which may be overwhelming for initial profiling, or the very specific Target 48 panels (focused only on cytokines), the Target 96 panels provide a curated set of biomarkers that is comprehensive yet manageable for a profiling study.
In summary, the most suitable panels are from the Target 96 series, as they offer an optimal balance of breadth, focus, and established biological relevance.
Here is a list of recommended panels we offer:
Table. List of Olink Panels
Additional Options
In addition to the panels above, we also offer the following panels:
- Explore HT
- Explore 3072 (3072 proteins)
- Explore 1536 (1536 proteins)
- Explore 384
- Target 48
- Custom Olink Flex systems
Our team can assist in designing an integrated Olink–Protein Biomarker Profiling strategy tailored to your project goals.
Applications of Protein Biomarker Profiling
Our Protein Biomarker Profiling Service is engineered to accelerate biomarker-driven insights, bridging genomic discoveries to functional protein-level understanding, it supports the following applications:
- Oncology Biomarker Discovery and Validation
Olink technology demonstrates particular strength in cancer biology research, where it facilitates the identification of specific protein signatures for the early recognition of biological changes, understanding disease progression, and monitoring dynamic biological responses. The technology's high sensitivity allows for the detection of low-abundance proteins that are critical for early biomarker discovery during phases when biological alterations are most discernible.
- Cardiovascular and Metabolic Disease Applications
In cardiovascular and metabolic disease research, Olink panels provide insights into pathophysiological processes and risk stratification. The technology's comprehensive inflammation coverage is particularly valuable, given the established role of inflammatory mechanisms in atherosclerosis and cardiovascular disease progression. Researchers can monitor dynamic protein expression changes across different disease stages, identifying biomarkers that predict progression and treatment response.
- Neurodegenerative and Mental Health Disorders
Olink proteomics enables investigation of protein networks underlying complex neurological and psychiatric conditions. For neurodegenerative conditions like Alzheimer's and Parkinson's disease, Olink's multiplex capabilities facilitate comprehensive profiling of proteins involved in disease pathogenesis, potentially leading to earlier diagnostic markers and therapeutic targets.
- Integrating Proteomics with Other Omics Data
Olink technology plays a pivotal role in multi-omics integration, particularly in connecting genomic discoveries with functional protein-level insights. This integrated approach is especially powerful for identifying causal protein biomarkers and validating therapeutic targets through proteogenomic analyses.
Workflow of Protein Biomarker Profiling
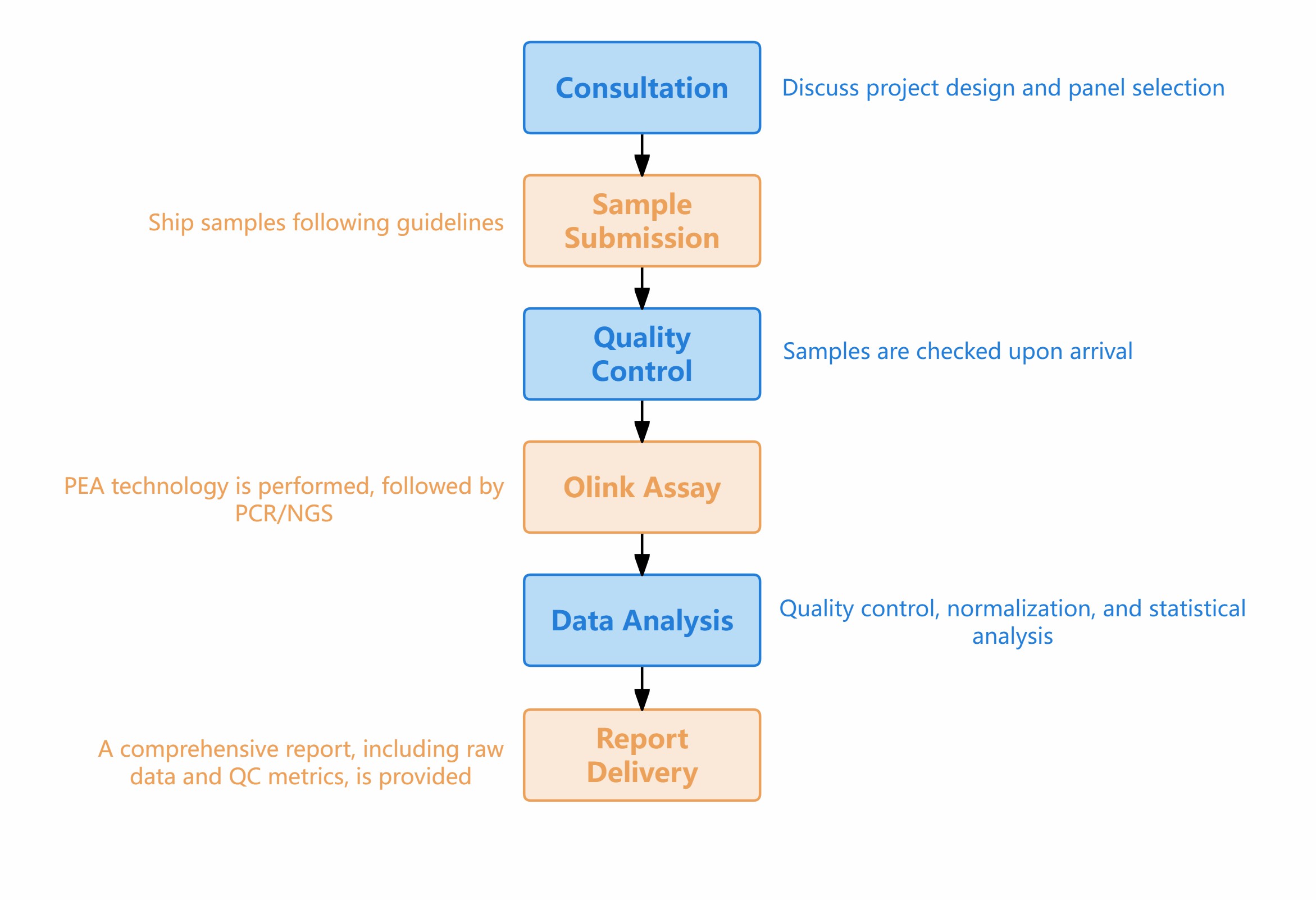 Figure 1: Workflow of Olink' s Protein Biomarker Profiling Service.
Figure 1: Workflow of Olink' s Protein Biomarker Profiling Service.
Why Creativ Proteomics
Advanced Analytical Technologies for Deep Profiling
The Olink technology achieves ultra-high sensitivity, detecting proteins at fg/mL levels, which is crucial for identifying low-abundance biomarkers in complex biological fluids like plasma or serum. The assay boasts a dynamic range spanning 10 orders of magnitude (log10), allowing for the simultaneous accurate quantification of high, medium, and low-abundance proteins in a single run without sample dilution.
High-Throughput and Comprehensive Coverage
The service enables the simultaneous profiling of a vast number of proteins from a single, minute sample volume. Panels are available for detecting 48 to over 5,400 proteins, with high-plex options like Explore HT (5,400+ proteins) and Explore 3072 (3,072 proteins) designed for large-scale discovery studies.
Robust Data Quality and Integration
Our service provides normalized protein expression data in Normalized Protein eXpression (NPX) units, which are log2-scaled for easy statistical analysis. This standardized output facilitates downstream bioinformatic analyses, such as differential expression, pathway analysis, and multi-omics integration.
Application-Oriented Service
We offer a complete service, from initial project consultation and sample collection guidance to standardized sample processing, quality-controlled Olink assays, data analysis, and detailed reporting. Our service is tailored for critical applications in biomedical research, including biomarker discovery and validation, drug target identification and mechanism-of-action studies, and multi-omics integration (e.g., proteogenomics).
Demo Results: Olink-Protein Biomarker Profiling
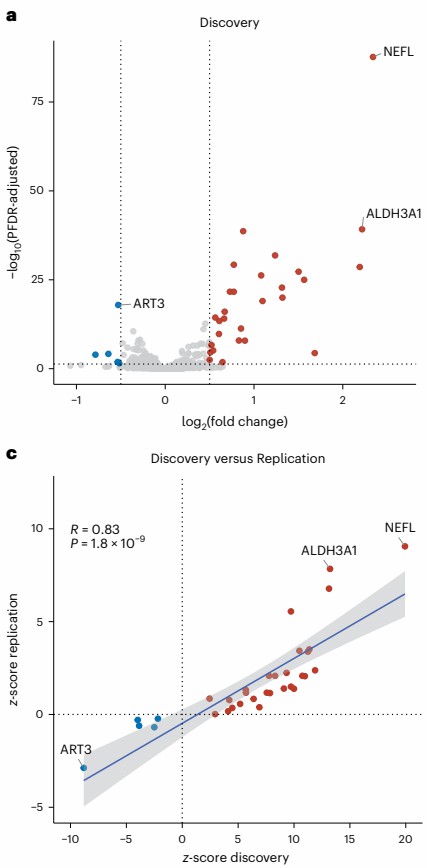 Figure 2: 33 differentially abundant plasma proteins in ALS patients versus controls in the Discovery Cohort. (Chia, R., et al. 2025)
Figure 2: 33 differentially abundant plasma proteins in ALS patients versus controls in the Discovery Cohort. (Chia, R., et al. 2025)
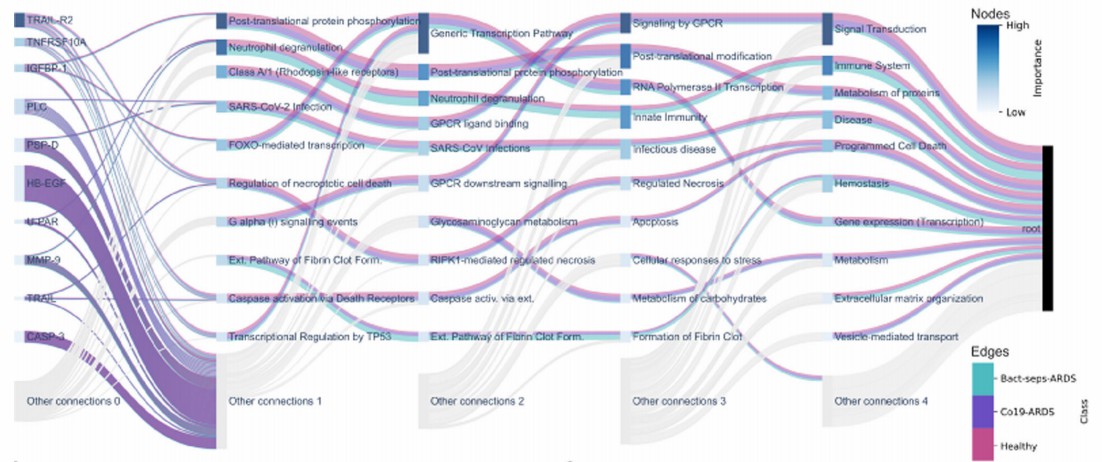 Figure 3: The BINN architecture trained on Olink proteomic data highlights nodes important for distinguishing COVID-19-induced ARDS, bacterial sepsis-induced ARDS, and healthy controls. (Hartman, E., et al. 2023)
Figure 3: The BINN architecture trained on Olink proteomic data highlights nodes important for distinguishing COVID-19-induced ARDS, bacterial sepsis-induced ARDS, and healthy controls. (Hartman, E., et al. 2023)
Sample Requirements for Protein Biomarker Profiling
1. Sample Types and Volumes
- Plasma/Serum (Recommended): Minimum 40-50 µL per sample is required.
- Other validated types: Cerebrospinal fluid, aqueous humor - 50 µL.
- Non-standard Sample Types (require pre-experiment validation): Cell culture supernatants, tissue lysates, cell lysates and exosome preparations.
2. Sample Storage and Shipping
- Storage temperature: -80°C.
- Shipping condition: On dry ice (samples must remain frozen upon arrival).
- Sample containers: Use temperature-resistant, non-protein-binding plasticware.
- Recommended: 96-well PCR plates with complete skirts and quality sealing membranes.
Case Study
Title: Comparative studies of 2168 plasma proteins measured by two affinity-based platforms in 4000 Chinese adults
Journal: Nat Commun
Year: 2025
- Background
- Methods
- Results
Proteomics provides critical insights into human biology and drug development, yet direct comparisons between affinity-based proteomic platforms remain limited. This study conducted a large-scale comparative analysis of plasma proteomic profiles measured by Olink Explore (antibody-based) and SomaScan (aptamer-based) technologies in 3,976 Chinese adults from the China Kadoorie Biobank. The aim was to evaluate the correlation between protein measurements, identify genetic variants influencing protein levels (pQTLs), assess associations with phenotypes like BMI and ischemic heart disease (IHD), and compare the predictive utility of both platforms for IHD risk.
The Olink Explore 3072 assay was used to measure plasma protein levels. For each participant, 40μL of plasma was aliquoted into 96-well plates, including external quality control (QC) samples. Paired antibodies targeting 2,923 unique proteins (mapped to UniProt IDs) were employed to bind specific protein epitopes. Protein levels were normalized using inter-plate controls and transformed into Normalized Protein eXpression (NPX) units on a log2 scale. The limit of detection (LOD) was defined using external QC samples, and sample-specific QC warnings were flagged based on deviations. Replicates across panels showed high correlations (r>0.8r > 0.8r>0.8), ensuring data reliability.
Olink demonstrated robust performance in protein quantification and genetic association analyses. The median observational correlation between Olink and SomaScan for 1,694 one-to-one matched proteins was modest (Spearman's ρ = 0.26), influenced by protein abundance and data quality parameters (e.g., % below LOD; Fig. 2a, b). Genome-wide association studies identified cis-pQTLs for 765 Olink proteins, with 400 showing colocalization with SomaScan cis-pQTLs (Fig. 2c, d). Phenotypically, 1,096 Olink proteins were significantly associated with BMI (FDR < 0.05), and 279 with IHD risk (Fig. 3a, d). Notably, adding Olink proteins to conventional risk factors improved IHD prediction, increasing the C-statistic from 0.845 to 0.862 (NRI: 12.2%; Fig. 4). Proteins with high observational correlations or colocalizing pQTLs (e.g., GHR for BMI, WFDC2 for IHD) showed consistent associations across platforms, underscoring Olink's reliability for biomarker discovery and risk prediction.
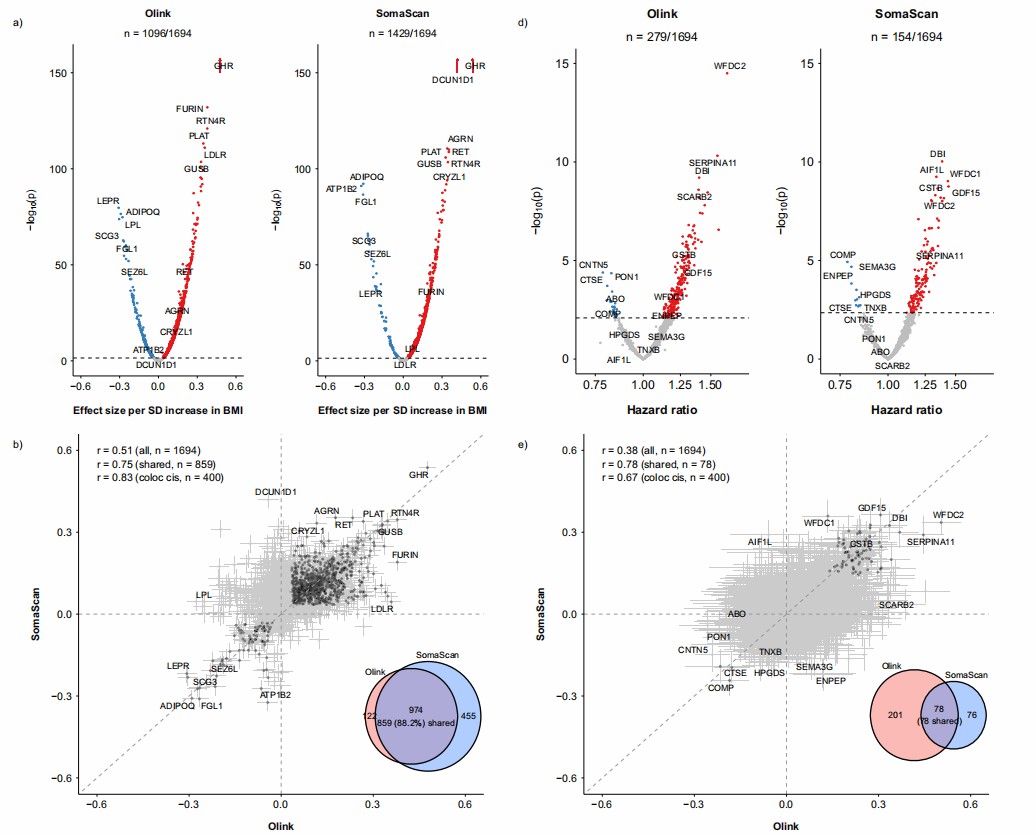 Figure 4: Comparison of number of proteins significantly associated with BMI and risk of incident IHD and their effect sizes between Olink and SomaScan platforms. (Chia, R., et al. 2025)
Figure 4: Comparison of number of proteins significantly associated with BMI and risk of incident IHD and their effect sizes between Olink and SomaScan platforms. (Chia, R., et al. 2025)
FAQs
Are there species-specific panels?
Yes. we offer Target 96 Mouse Exploratory and Target 48 Mouse Cytokine panels. While human-focused panels are designed for human samples, some antibodies may show cross-reactivity with homologous proteins in other species.
How many replicates are needed?
Technical replicates are typically not required due to the low technical variation (most assays show intra-assay CVs <10%). Biological replication (using different samples) is generally more critical.
Can I compare results from different Olink panels?
While absolute NPX values may differ between panels, the relative expression ratios for a given protein across samples are expected to be consistent.
References
- Chia, R., Moaddel, R., Kwan, J.Y. et al. A plasma proteomics-based candidate biomarker panel predictive of amyotrophic lateral sclerosis. Nat Med (2025).
- Hartman, E., Scott, A.M., Karlsson, C. et al. Interpreting biologically informed neural networks for enhanced proteomic biomarker discovery and pathway analysis. Nat Commun 14, 5359 (2023).
- Wang, B., Pozarickij, A., Mazidi, M. et al. Comparative studies of 2168 plasma proteins measured by two affinity-based platforms in 4000 Chinese adults. Nat Commun 16, 1869 (2025).