Introduction: Why Alzheimer's Research Needs Better Tools
Alzheimer's disease (AD) continues to challenge research teams because its biological changes begin many years before clinical symptoms appear. Traditional biomarkers—amyloid‐β, tau proteins, neuroimaging—offer powerful insights, but they have limits: high cost, invasive sampling (e.g. lumbar puncture), and difficulty capturing the full molecular complexity of neurodegeneration. To make progress in early detection, patient stratification, and mechanism discovery, researchers need tools that are sensitive, multiplexed, and usable in large cohorts.
That's where Olink proteomics panels, especially Olink serum proteomics using proximity extension assay (PEA) technology, offer real value. These allow simultaneous measurement of dozens to thousands of proteins from small serum or plasma samples, with high sensitivity for low‐abundance molecules. In Alzheimer's research, application of Olink panels is helping to reveal novel protein signatures linked to inflammation, synaptic loss, vascular changes, and other pathological processes beyond amyloid and tau.
In this article, we examine how researchers are using Olink panels in Alzheimer's disease research: what advantages these assays bring, what real case‐studies show, and how they might accelerate non‐clinical discovery projects. (Note: all applications discussed are for research use. Not clinical diagnosis or treatment.)
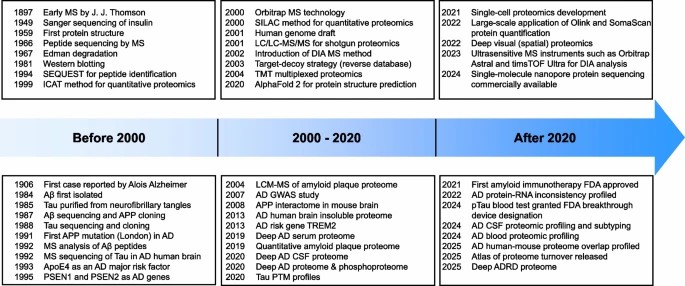 Fig 1.Evolution of proteomics methodologies in AD research.
Fig 1.Evolution of proteomics methodologies in AD research.
Why Olink Panels Matter in Neurodegeneration
When studying Alzheimer's or related neurodegenerative disorders, researchers face serious challenges: proteins in blood are often low-abundance; disease mechanisms involve many pathways (inflammation, synaptic loss, vascular change); reproducibility across labs and cohorts is often weak. Olink panels (especially serum/plasma proteomics via PEA) help address these exact obstacles.
Technical Advantages of Olink Proteomics
High sensitivity and specificity
Olink's PEA technology uses matched antibody pairs plus DNA-oligonucleotides. Only when both antibodies bind the same target does a DNA extension event occur. That reduces cross-reactivity and background signal.
Low sample volume required
Many Olink panels need as little as ~1-3 µL serum or plasma. This is essential when sample amounts are limited (e.g. in longitudinal Alzheimer's studies, rare dementias, or small animal models).
Wide dynamic range and multiplexing
Olink panels cover proteins spanning many orders of magnitude in abundance. They allow simultaneous interrogation of many proteins (from 48/96-plex up to high-plex or full library formats) in one assay.
Reproducibility and biotemporal stability
Studies show that many analytes measured by Olink in plasma show good intra- and inter-plate precision. Analyses of sample stability over time (within individuals) suggest that protein measurements remain relatively stable, allowing meaningful longitudinal studies.
Scalability from discovery to validation
One can use high-plex Olink panels for discovery, then move to mid- or low-plex or even custom panels for validation or signature development without changing the core assay technology. This reduces technical drift and confounders when comparing across cohorts.
Application Advantages for Alzheimer's / Neurodegenerative Research
Capturing multiple pathological pathways simultaneously
Alzheimer's involves more than amyloid-β and tau. Inflammation, synaptic dysfunction, vascular injury, oxidative stress all contribute. Olink profiling permits observation of many of these pathways in parallel. For example, in a multi-platform proteomics study comparing CSF and plasma, Olink detected co-expression modules related to proteostasis and matrisome in AD, correlating with known brain pathology.
Potential for less invasive biomarker sources
CSF is informative but collection is invasive. Plasma/serum is far easier to collect repeatedly. Olink serum proteomics has been used to detect Alzheimer's-associated dysregulation of proteins in blood, opening possibilities for large cohort screening or monitoring.
Translatability across cohorts and populations
High reproducibility and standardisation in panels means observed signatures are more likely to be validated across different populations, reducing bias due to demographic or technical variation. For example, the large-scale plasma proteomic profiling using Olink Target 96 was validated in independent cohorts, achieving high performance in distinguishing Alzheimer's patients across populations.
Facilitating signature / biomarker panel development
Because Olink allows discovery and validation within similar technological frameworks, signatures of Alzheimer's disease stage, or of disease vs control, can be developed more efficiently. For example, a 19-protein signature was identified in plasma in one cohort to classify AD vs controls with very high AUC.
Limitations & Considerations (for Researchers to Keep in Mind)
While powerful, Olink panels are not a panacea. Good research design must account for:
Multiple testing burden
With high-plex panels, correction for multiple comparisons reduces statistical power. Large sample sizes are often needed.
Biological variability over time
Even in healthy individuals, some protein levels fluctuate. Researchers must consider intra-individual (biotemporal) variability when planning longitudinal studies.
Matrix effects and sample handling
Differences in sample collection, storage, handling (e.g., freeze-thaw cycles), and choice of serum vs plasma vs CSF can affect results. Standard protocols are essential. (While Olink protocols are strong, such preanalytical variation remains a concern.)
Relative quantification vs absolute
Many Olink panels report relative expression (NPX units) rather than absolute concentration, which may limit cross-study comparability unless panels calibrated or custom absolute units used.
Case Study: Olink Serum Proteomics in Alzheimer's Cohorts
Here we highlight several peer-reviewed Alzheimer's disease (AD) research projects that have successfully used Olink serum/plasma proteomics. These illustrate how Olink panels are applied, what signatures emerge, and what pitfalls to watch out for.
Study 1: Swedish BioFINDER — Early Alzheimer's Changes in Plasma & CSF
Design & Cohorts
The study used the Olink Target 96 / ProSeek platform to assay ~270 proteins in cerebrospinal fluid (CSF) and plasma across large groups: cognitively normal participants, mild cognitive impairment (MCI), and AD patients, stratified by amyloid-β (Aβ) status.
A validation cohort was included to confirm findings.
Main Findings
Ten CSF proteins (e.g. CHIT1, SMOC2, MMP-10, LDLR, CD200, EIF4EBP1, ALCAM, RGMB, tPA, STAMBP) differed in Aβ+ individuals vs controls.
Six plasma proteins (OSM, MMP-9, HAGH, CD200, AXIN1, uPA) showed altered levels in Aβ+ individuals.
Some analytes correlated strongly with cognitive performance and cortical thickness metrics.
The plasma signature achieved high diagnostic discrimination: AUC ~0.94 for distinguishing AD dementia vs controls, ~0.78 for prodromal AD vs controls.
Implications
This study demonstrates Olink serum/plasma proteomics can detect changes early (pre-symptomatic or prodromal), not just in dementia stage. It shows how immune/inflammation-related proteins, vascular/remodelling markers, and matrix/adhesion proteins are involved beyond classic amyloid/tau pathology.
Study 2: Technical Performance in Longitudinal Cohort
Design & Goal
To test reproducibility, stability, and variance over time of Olink PEA measurements. Plasma samples collected annually for three years from both AD and cognitively unimpaired subjects (n = 54 total). Multiple Olink 96 panels were used, including Neuro exploratory, Inflammation, Cardiometabolic, etc.
Key Findings
Most measured analytes had good technical precision (intra- and inter-plate), showing the assay is robust for longitudinal studies.
Biotemporal stability (variation of same-person over time) was moderate: many proteins were stable, others varied more, influenced by biological and individual factors.
Variance component analysis showed that for many proteins, inter-individual differences (diagnosis, age, etc.) exceed within-person noise. This suggests that meaningful biological signal is accessible, provided sample size and design are sufficient.
Take-home
For researchers planning longitudinal Alzheimer's biomarker studies, this shows Olink serum/proteomics panels are technically feasible. However, sample handling, choice of panels, and statistical power (to correct for multiple comparisons) are critical.
Study 3: Plasma Biomarker Panel from Hong Kong Chinese Cohort
Design
Using Olink Target 96 panels to assay ~1,160 plasma proteins in Alzheimer's disease (AD) vs healthy controls in a Chinese cohort, with validation in independent sample sets.
Results
Found ~429 proteins dysregulated in AD plasma.
Developed a 19-protein "hub" signature that classifies AD vs controls with high accuracy (AUC 0.97-0.98).
Some of the hub proteins' expression varied depending on disease stage, which aids in staging or tracking progression.
Significance
This shows that a focused proteomic signature, when derived from a large multiplex Olink panel, can perform well in discriminating AD in non-CSF fluid. Also, it shows generalizability when validated in independent cohorts.
Study 4: Early-Stage AD & Mild Cognitive Impairment — Inflammation Panel (China)
Design
30 individuals total: 10 healthy controls, 10 PET-confirmed AD, 10 MCI (PET negative), using Olink inflammation panel (PEA) to assay ~92 inflammation-related proteins in plasma.
Key Findings
Several proteins (uPA, CX3CL1, CDCP1, Flt3L, SCF, TWEAK) were elevated in plasma of AD vs controls. Some also elevated in MCI.
ROC analysis showed excellent discrimination: e.g., uPA had AUC ≈ 0.96.
Functional enrichment pointed to neuroinflammation, synaptic dysfunction, and maybe vascular involvement.
Lessons
Even with modest samples, targeted inflammation-panel Olink assays can identify differentially expressed proteins in early AD/MCI, suggesting possible biomarkers or mechanistic pathways. But small cohort size limits generalizability; requires further validation.
Summary & Comparative Insights
| Aspect | Strengths Demonstrated | Common Challenges |
| Diagnostic discrimination | High AUCs (≥ 0.94) in distinguishing AD vs controls in some studies. | Lower performance for prodromal stages; overlapping signals. |
| Early-stage detection / preclinical changes | Swedish BioFINDER & Hong Kong studies show changes before overt symptoms. | Need for large, longitudinal cohorts to validate stage-specific biomarkers. |
| Panel depth & signature derivation | Multiplex assays (hundreds-to-thousands of proteins) allow discovery of large dysregulated sets and focused hub signatures. | Managing multiple testing & false discovery; handling sample handling & variability. |
| Reproducibility & stability | Longitudinal technical performance is good; many proteins stable over time. | Some analytes have high intra-individual variance; need rigorous QC, sample handling, power. |
Translational Impact: From Biomarkers to Drug Development
While Olink serum/plasma proteomics is not used for diagnosis or treatment, it has strong potential to inform early-stage drug development, help select targets, stratify preclinical models, and monitor pharmacodynamics in non-clinical studies. Below are concrete ways that findings in Alzheimer's research translate, plus examples of environments where this is already happening.
Using Olink Panels to Prioritise Drug Targets
Mendelian Randomization + Proteomics for Causal Inference
A recent study integrated plasma proteomics with pQTL (protein quantitative trait loci) data and GWAS (genome-wide association study) summary statistics. They identified five proteins (BLNK, CD2AP, GRN, PILRA, PILRB) whose genetically predicted levels are likely causally associated with Alzheimer's disease risk. Of these, GRN (progranulin) appeared protective, others as risk factors. This kind of data helps drug developers pick proteins anchored in genetic evidence. (Zhan et al., 2025, J Transl Med. DOI:10.1186/s12967-025-06317-5)
Inflammation Markers as Therapeutic Leverage Points
In an Olink proteomics study of early-stage Alzheimer's / mild cognitive impairment, several inflammation-related proteins were elevated (for example uPA, CX3CL1, CDCP1, TWEAK, etc.), with ROC curves showing good discrimination. These proteins, especially ones consistently altered, can be considered for therapeutic modulation or for use as biomarkers of pharmacodynamic response in non-clinical trials. (Wei et al., 2025. DOI:10.3389/fneur.2025.1615152)
Enabling Better Preclinical & Translational Study Design
Signature Panels Guide Model Validation
With Olink panels, the same biomarkers identified in human Alzheimer's cohorts can be checked in animal or cellular disease models. If the protein signature or pathway perturbation is recapitulated in a model, it strengthens the relevance of that model for testing drug candidates.
Stratification & Patient / Sample Selection in Early-Stage Studies
Proteomic signatures from Olink allow grouping by molecular subtype even among clinically similar subjects. For example, distinguishing people by inflammatory profiles or vascular injury signatures could allow non-clinical or ex vivo studies to be more focused, and preclinical efficacy studies to be powered more efficiently.
Monitoring Pharmacodynamics & Biomarker Response in Non-Clinical Studies
Use in Early Safety / Mechanism-of-Action Studies
Olink's capacity to monitor many serum (or plasma) proteins with small volume allows measuring the downstream effect of candidate drugs in animals or in cell culture (e.g. secreted proteins). If a drug is supposed to affect inflammation, synaptic function, or vascular remodeling, then Olink panels can monitor relevant protein changes over time.
Biomarker-led Go/No-Go Decisions
In drug development pipelines, using Olink proteomics as a readout in non-clinical toxicology or pharmacology sections helps make early decisions: if the expected biomarker signature is not modulated, it may signal insufficient target engagement or off-target effects—allowing resource shift early.
Examples of Research Toward Translational Practice
Multiplex Proteomics Identifies Novel CSF and Plasma Biomarkers
The Swedish BioFINDER cohort (Whelan et al., 2019) used Olink Target 96 panels to identify proteins in both CSF and plasma that are altered in Alzheimer's (versus controls), including those associated with immune response, angiogenesis, and matrix remodelling. Such markers offer pathways that might be druggable, or at least monitorable.
Proteomic Landscape Review: Mechanism + Biomarker Discovery
A recent review (Molecular Neurodegeneration, 2025) summarises how affinity-based platforms (like Olink) alongside mass spec are revealing not just biomarkers but mechanistic insight—protein turnover, matrisome / extracellular matrix changes, proteostasis, etc.—which are central to identifying new drug target hypotheses.
Key Considerations for Translational Use
Validation & Cross-Cohort Reproducibility
Any biomarker or target from an Olink study should be validated in independent cohorts (different populations, geographies). This ensures robustness. Many Alzheimer's biomarker findings suffer from overfitting, small sample size, or cohort effects.
Relative vs Absolute Quantification
Olink typically gives relative expression (NPX units). For translational or regulatory preclinical studies, there may be a need to establish absolute quantification or calibrate assays for comparability.
Standardised Sample Handling & Timing
Preanalytical variables (collection tube type, time to freeze, multiple freeze-thaw cycles) can introduce noise. For use in drug development contexts, strict SOPs are needed.
Biological Variability & Confounders
Age, sex, comorbidities (e.g. vascular disease), medications all impact proteomics signatures. They must be modelled or stratified in study design.
Conclusion
The use of Olink serum/plasma proteomics is opening new frontiers in Alzheimer's disease research. As we've seen, Olink panels overcome many historical limitations—enabling multiplexed, high-sensitivity measurement of proteins from small sample volumes, capturing multiple pathological pathways beyond amyloid and tau, and documenting changes even in early or prodromal stages. Case studies from BioFINDER, Hong Kong cohorts, and longitudinal work confirm that serum signatures correlate with cognition, imaging, and disease progression, and that many markers remain stable enough over time to support translational objectives.
Looking ahead, several directions will sharpen impact for non-clinical R&D teams and academic researchers:
Emerging Trends & Opportunities
Affinity-based + Mass Spectrometry Hybrid Approaches
Recent reviews show that combining Olink (and other affinity-based platforms) with MS-based proteomics reveals richer networks of altered proteins, especially in biofluids. These hybrid designs help cross-validate findings and identify proteins that might be missed by one technology alone. (Yarbro et al., Molecular Neurodegeneration, 2025)
Multi-cohort & Multi-biofluid Designs
Studies increasingly use matched CSF/plasma samples, or multiple independent cohorts, to confirm that findings aren't cohort or sample-handling specific. This is key for reproducibility. The Multi-platform proteomics analyses confirm that some proteins show consistent directionality across fluids and platforms.
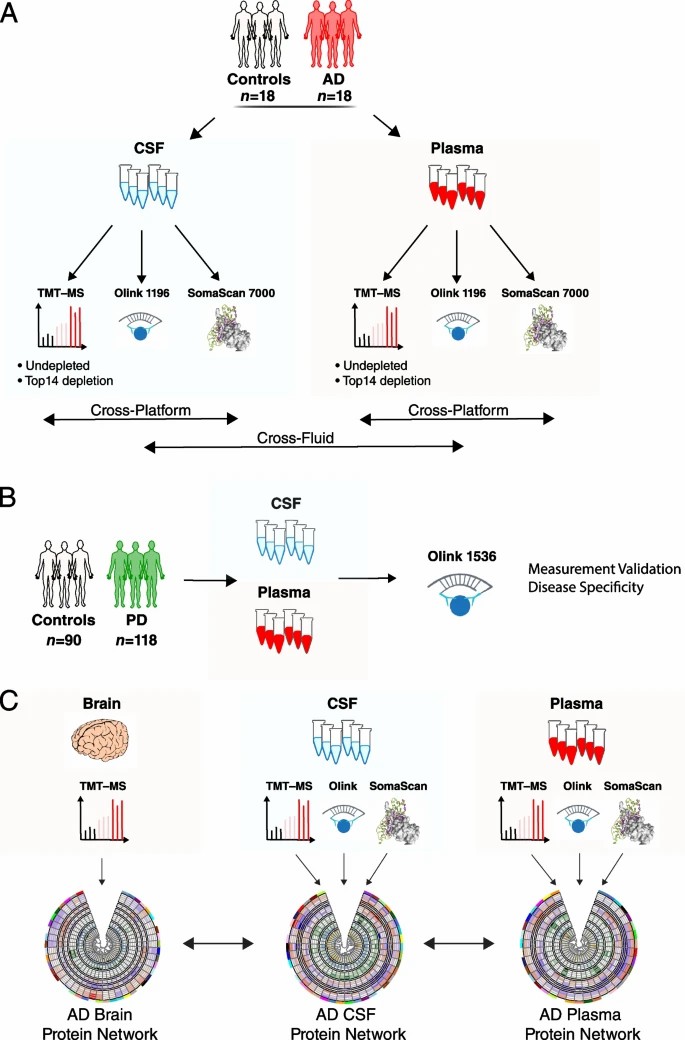 Fig 2. Multi-platform proteomics analyse Study overview.
Fig 2. Multi-platform proteomics analyse Study overview.
Inflammation and Subtypes
Inflammation researchers using Olink panels are observing that plasma inflammatory markers not only differ in Alzheimer's vs controls, but also help define subtypes (e.g. high inflammation vs low). Understanding these subtypes can improve model systems and drug target selection.
Integration with Genetics, Imaging, and Other Omics
Linking proteomic signatures with genetic risk (e.g. pQTL), imaging biomarkers, and transcriptomics is delivering insights into causal proteins and disease mechanisms. Recent efforts have identified consensus altered proteins like SMOC1, VGF, NRN1, etc., through this integrative lens.
Custom Panels & Focused Signatures
As findings solidify, there's growing interest in moving from discovery (hundreds-to-thousands of proteins) to more focused custom panels. These are more cost-effective, require less sample, and are better suited to repeated or high-throughput non-clinical studies. The Amsterdam UMC/Teunissen et al. custom panels are examples.
Practical Next Steps for Research Teams & CROs
Define study design up front to include:
- adequate sample size for early vs later disease stages
- matched sample types (e.g. serum/plasma, or CSF/plasma if possible)
- strict sample handling SOPs to reduce pre-analytical variation
- Prioritize cross-cohort validation: after initial discovery in one population, validate in a separate cohort (geography, ethnicity, sample type) to ensure signals hold up.
- Use hybrid technologies when possible: combining Olink with MS or other platforms strengthens confidence in biomarker candidates.
- Plan assays for translation: once potential biomarkers are found, consider how they will be used in downstream non-clinical studies (mechanism of action, target engagement, safety), and whether relative quantification is sufficient or absolute quantification is needed.
- Collaborate early with CROs / biotech / academic partners to align biomarker goals, access shared sample resources, and ensure reproducibility.
If your lab or organisation is exploring proteomic biomarkers for Alzheimer's but not yet using Olink panels, here's how to get started:
- Reach out to our Olink proteomics team to discuss which panel(s) fit your Alzheimer's-oriented projects.
- Request a pilot study to test serum proteomics in your cohort — this helps calibrate sample handling, variance, and biomarker performance.
- We believe that with rigorous design, technology integration, and thoughtful validation, Olink serum proteomics can drive major non-clinical advances in Alzheimer's biomarker discovery, target prioritization, and translational research. Let's build together.
References
- Zhan, H., Cammann, D., Cummings, J.L. et al. Biomarker identification for Alzheimer's disease through integration of comprehensive Mendelian randomization and proteomics data. J Transl Med 23, 278 (2025).
- Yarbro, J.M., Shrestha, H.K., Wang, Z. et al. Proteomic landscape of Alzheimer's disease: emerging technologies, advances and insights (2021 – 2025). Mol Neurodegeneration 20, 83 (2025).
- Dammer, E.B., Ping, L., Duong, D.M. et al. Multi-platform proteomic analysis of Alzheimer's disease cerebrospinal fluid and plasma reveals network biomarkers associated with proteostasis and the matrisome. Alz Res Therapy 14, 174 (2022).
- Wei J, Ni H, Liu W, Xu H, Wang C, Geng G, He Z, Chen G. Biomarkers and therapeutic targets in early Alzheimer's disease: an Olink proteomics study. Front Neurol. 2025 Jul 17;16:1615152. doi: 10.3389/fneur.2025.1615152. PMID: 40746641; PMCID: PMC12310730.
- Dammer, E.B., Ping, L., Duong, D.M. et al. Multi-platform proteomic analysis of Alzheimer's disease cerebrospinal fluid and plasma reveals network biomarkers associated with proteostasis and the matrisome. Alz Res Therapy 14, 174 (2022). https://doi.org/10.1186/s13195-022-01113-5
- Jiang Y, Zhou X, Ip FC, Chan P, Chen Y, Lai NCH, Cheung K, Lo RMN, Tong EPS, Wong BWY, Chan ALT, Mok VCT, Kwok TCY, Mok KY, Hardy J, Zetterberg H, Fu AKY, Ip NY. Large-scale plasma proteomic profiling identifies a high-performance biomarker panel for Alzheimer's disease screening and staging. Alzheimers Dement. 2022 Jan;18(1):88-102. doi: 10.1002/alz.12369. Epub 2021 May 25. PMID: 34032364; PMCID: PMC9292367. https://doi.org/10.1002/alz.12369
- Carlyle BC, Kitchen RR, Mattingly Z, Celia AM, Trombetta BA, Das S, Hyman BT, Kivisäkk P, Arnold SE. Technical Performance Evaluation of Olink Proximity Extension Assay for Blood-Based Biomarker Discovery in Longitudinal Studies of Alzheimer's Disease. Front Neurol. 2022 Jun 6;13:889647. doi: 10.3389/fneur.2022.889647. PMID: 35734478; PMCID: PMC9207419. https://doi.org/10.3389/fneur.2022.889647
- Whelan, C.D., Mattsson, N., Nagle, M.W. et al. Multiplex proteomics identifies novel CSF and plasma biomarkers of early Alzheimer's disease. acta neuropathol commun 7, 169 (2019). https://doi.org/10.1186/s40478-019-0795-2








