- Overview
- Panels List
- Applications
- Workflow
- Why Creative Proteomics
- Demo
- Sample Requirements
- Case
- FAQ
- Sample Submission Pack
Customized and Ready-to-Use Olink Proteomics Panel for Plasma &Serum Sample
Why Analyze Blood Samples?
1. Non-Invasive Sampling – Plasma and serum are easily accessible biofluids, enabling large-scale studies without complex procedures.
2. Systemic Disease Insights – Reflect real-time physiological and pathological changes, making them ideal for studying inflammation, infection, and chronic diseases.
3. Dynamic Protein Profiles – Capture circulating proteins, signaling molecules, and metabolic markers missed in tissue-specific analyses.
By focusing on plasma & serum proteomics, researchers gain a direct window into systemic biology—unlocking actionable biomarkers with clinical relevance.
What We Provide
Our service utilizes Olink's proximity extension assay (PEA) technology to deliver sensitive, high-throughput protein quantification.
- Minimal sample required: As little as 1 µL of plasma or serum
- High sensitivity: Detects proteins in the low pg/mL range
- Excellent reproducibility: Coefficient of variation (CV) <10%
Researchers can choose from Olink's pre-validated panels or fully customize protein targets based on specific biological pathways or research needs.
This solution is ideal for:
- Biomarker discovery
- Immune and inflammatory profiling
- Disease mechanism studies (e.g., oncology, cardiometabolism, neurology)
- Ready-to-use, scalable, and tailored to accelerate your proteomics research.
Features of the panel
- Species : Optimized for human proteome analysis.
- Design : Select target proteins via intuitive bioinformatics interface.
- Proteins : enabling targeted 21-plex to 384+ plex panels for cytokine, immune response, and pathway-specific research.
- Sample : 1 µL serum/plasma per analysis.
- Readout : NPX-based protein measurement.
- Platform : Proprietary PEA technology on Signature Q100.
List of Our Plasma & Serum Proteome Assay Panel
Protein category
The Plasma & Serum Proteome Assay analyzes proteins across multiple functional categories essential for biological research, including Metabolic Enzymes, Signaling Molecules, Immune Regulators, Extracellular Matrix Components, Hormones/Binding Proteins, and Stress Response Factors (see Table. List of Plasma & Serum Proteome Assay).
The assay covers key biological processes such as cellular communication, metabolic regulation, inflammatory pathways, and tissue remodeling, featuring carefully selected targets implicated in fundamental physiological mechanisms. Through rigorous literature curation, we incorporate scientifically validated biomarkers associated with metabolic networks, cellular homeostasis, and systemic regulation, providing researchers with a comprehensive tool for proteome-wide investigations across diverse research areas including metabolic studies, aging research, and cellular response mechanisms. The panel's systematic organization enables targeted analysis of protein networks while maintaining the depth required for mechanistic discovery research.
Table. List of Plasma & Serum Proteome Assay
Protein Functions
Biological process
primarily involved in key biological processes including metabolic regulation, cellular signaling pathways, stress response mechanisms, immune system modulation, and tissue homeostasis maintenance.
Disease area
primarily associated with research areas involving metabolic disorders, chronic inflammatory states, age-related pathological processes, neoplastic transformations, and neurological degeneration models.
The Application of Plasma &Serum Proteome Assay.
The Plasma & Serum Proteome Assay enables high-throughput quantification of protein biomarkers across biological pathways, providing researchers with a powerful tool for:
- Discovery of protein networks underlying metabolic dysregulation in aging models and nutritional intervention studies;
- Mechanistic investigation of stress response pathways in cellular senescence and oxidative damage research;
- Identification of molecular signatures associated with tissue remodeling in experimental inflammatory models.
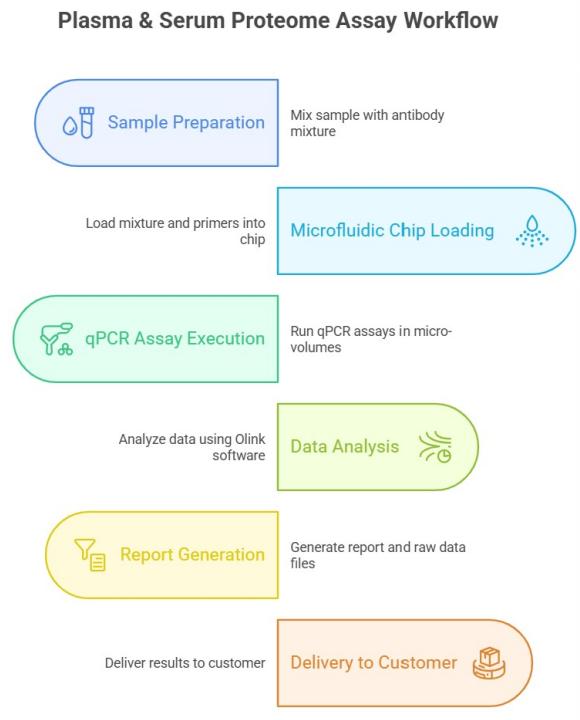
Workflow of Olink Proteomics
Why Creativ Proteomics
Pathway-Specific Analysis
Preconfigured bioinformatics workflows for metabolic, inflammatory, and stress-response pathways with 50+ validated biomarker ratios.
Customizable Panel Design
Flexible configuration with 30+ adjustable targets complementing 400+ core protein measurements for study-specific needs.
Comprehensive Protein Coverage
Simultaneous measurement of 500+ proteins spanning multiple functional categories (signaling, immune regulation, metabolism).
Advanced Bioinformatics
Built-in tools for pathway enrichment, network analysis, and machine learning-ready data formats.
Demo Results of Olink Data
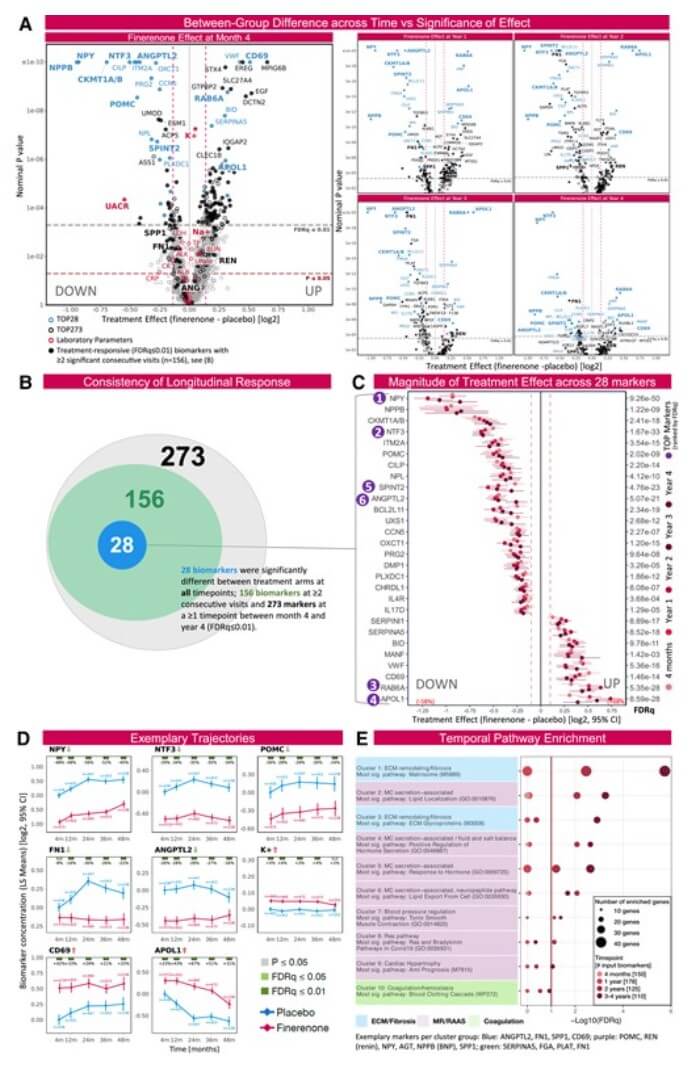 Figure 1: Finerenone induces sustained biomarker changes while potentially influencing remodeling and RaaS pathways. (Berger, M., et al. 2025)
Figure 1: Finerenone induces sustained biomarker changes while potentially influencing remodeling and RaaS pathways. (Berger, M., et al. 2025)
Sample Requirements
| Sample Type | Recommended Sample Size | Sample Quality | Pre-treatment and Storage | Sample Transport |
| Plasma/Serum/Body Fluid | 40 µL minimum sample volume required per analysis. | Optimal Concentration: 0.5–1 mg/mL | Storage Protocol: Aliquot and store at -80°C | Dry ice shipment at -80°C for sealed samples |
| Tissue | ||||
| Cells | ||||
| Exosomes | ||||
| Other |
Case Study

Comparing Venous vs. Capillary Blood Collection Methods for Proteomic Measurement in Peripheral Blood
Journal: Proteomics. Clinical applications
Year :2025
- Background
- Methods
- Results
Capillary blood sampling offers significant benefits compared to venous blood collection, particularly with the growing emphasis on decentralized clinical trials and field testing. However, no existing research has evaluated the performance differences between these two sampling methods for proteomic analysis. This study aims to assess the comparability of venous and capillary blood collection techniques in peripheral blood proteomics.
The Olink Explore 384 inflammatory panel was used to analyze 368 proteins in paired venous and capillary blood samples from 22 participants. Protein expression levels were assessed using descriptive statistics and correlation analysis. Additionally, we focused on a subset of proteins previously linked to morbidity and mortality in recent studies.
Of the 3 proteins detected, 2 had a strong positive correlation (r≥0.7) between protein concentrations determined in venous and capillary blood samples (215 of 327,66%). Another 47 cases (14%) had a moderate positive correlation (0.4≤r <0.7), and the remaining 65 cases (20%) had a weak or very weak correlation (r < 0.4). In capillary blood samples, protein expression was consistently high, but the correlation between different sampling methods was low (r < 0.6). We need further research to understand the potential reasons for the persistent low expression of proteins detected in venous samples/persistently high expression of a few proteins detected in capillary samples.
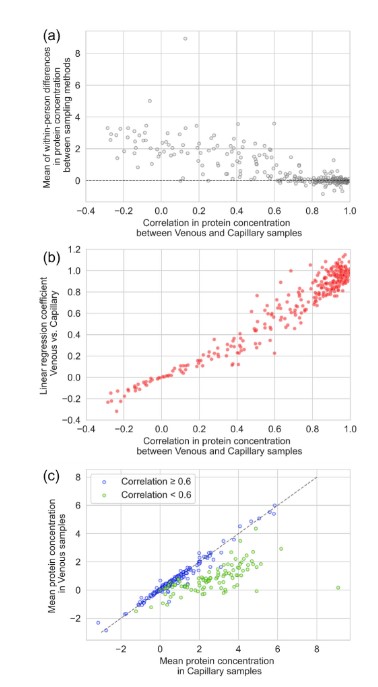 Figure 1. Intra-individual differences in protein concentrations between venous and capillary blood samples. ( Ni Lochlainn, M., et al. 2025)
Figure 1. Intra-individual differences in protein concentrations between venous and capillary blood samples. ( Ni Lochlainn, M., et al. 2025)
FAQs
What types of research applications is this platform designed for?
The technology supports diverse fundamental investigations including but not limited to mechanistic studies of cellular stress responses, comprehensive profiling of metabolic pathway alterations, systems-level analysis of immune modulation networks, biomarker discovery for physiological adaptations, and molecular phenotyping in experimental model systems.
How extensive are the protein target customization options?
Researchers can configure tailored panels ranging from focused 25-plex assays to comprehensive 1000+ protein profiles, selecting from our validated library of over 3,500 human protein targets with optional additions of synthetic peptide standards, internal reference controls, and study-specific normalization sets while maintaining the platform's stringent sensitivity and reproducibility standards.
What are the sample requirements and handling considerations?
The assay requires minimal input volumes (1 μL plasma/serum) and demonstrates consistent performance across various collection tubes (EDTA/heparin plasma, serum), with validated stability through three freeze-thaw cycles and recommended storage at -80°C with detailed pre-analytical variable documentation provided for experimental planning.
How does the assay facilitate multi-omics integration?
The platform provides built-in compatibility with transcriptomic and metabolomic data through unified file formats, coupled with specialized software tools for cross-omics pathway mapping, co-expression network analysis, and multidimensional data visualization, enabling true systems biology research approaches.
References
- Berger, M., MacNamara, A., Ferreira, J. P., et al. (2025). Finerenone effects on biomarkers: an analysis from the FIGARO-DKD trial. European heart journal, ehaf316. Advance online publication. https://doi.org/10.1093/eurheartj/ehaf316
- Ni Lochlainn, M., Cheetham, N. J., Falchi, M., et al. (2025). Comparing Venous vs. Capillary Blood Collection Methods for Proteomic Measurement in Peripheral Blood. Proteomics. Clinical applications, 19(4), e70007. https://doi.org/10.1002/prca.70007