Protein biomarker analysis enables researchers to identify and measure proteins that signal disease presence or progression. The global market for cancer biomarkers reached USD 38.14 billion in 2024, with rapid growth expected. Multiplex biomarker panels and advanced proteomics technologies, such as olink proteomics multiplex biomarker panels, improve detection rates by allowing simultaneous analysis of thousands of proteins. High-throughput platforms like Olink empower scientists to explore complex biological samples efficiently, accelerating biomarker discovery in cancer research.
Key Takeaways
- Protein biomarkers are crucial for detecting diseases and monitoring health changes.
- Multiplex biomarker panels allow simultaneous analysis of thousands of proteins, enhancing cancer research.
- High-throughput platforms like Olink improve sensitivity and efficiency in biomarker discovery.
- Understanding the types of protein biomarkers helps researchers design effective experiments.
- Selecting the right detection method is essential for achieving reliable results in cancer studies.
- High-sensitivity assays can detect low-abundance proteins, which are vital for early cancer detection.
- Integrating multi-omics data enhances the understanding of disease mechanisms and biomarker relevance.
- AI-driven approaches are transforming biomarker discovery by identifying patterns in complex datasets.

Introduction
What Are Protein Biomarkers?
Protein biomarkers are molecules that provide measurable information about biological processes. Scientists use these proteins to understand what happens inside cells and tissues. Protein biomarkers can indicate normal functions, disease states, or responses to treatments. Researchers often measure these proteins to track changes in health or disease.
- Protein biomarkers can signal the presence of disease or monitor how a condition changes over time.
- They include a wide range of biological indicators, such as proteins found in blood, tissue, or other body fluids.
- Scientists define a biomarker as a characteristic that can be measured objectively to show normal or abnormal biological activity.
Importance of Protein Biomarker Analysis in Research and Cancer Screening
Protein biomarker analysis plays a key role in modern research. Scientists rely on these measurements to predict disease risk and identify early signs of cancer.
- Protein biomarkers help researchers select high-risk groups for further study.
- They support decisions about which samples to investigate more closely.
- Researchers use protein biomarkers to monitor how diseases progress and how treatments work.
In cancer research, protein biomarkers have become essential tools. They help identify which proteins change during cancer development. Scientists have found that proteins often drive processes like tissue growth and cell division. By analyzing these proteins, researchers can discover new ways to detect cancer earlier.
Advances in technology, such as olink proteomics multiplex biomarker panels, allow scientists to measure thousands of proteins at once. This high-throughput approach improves the chances of finding important biomarkers. It also helps researchers understand complex biological pathways involved in cancer.
Purpose of This Resource Article
This article aims to provide a comprehensive overview of protein biomarker analysis for research professionals. Readers will learn about the main types of protein biomarkers, their roles in disease, and the latest detection methods. The article will compare traditional and advanced technologies, including olink proteomics multiplex biomarker panels, to highlight their strengths in cancer screening and discovery.
Note: This guide focuses on technical principles, workflows, and research applications. It does not address clinical or patient-oriented topics.
By the end of this article, readers will understand how to choose the right detection method for their research goals and how high-throughput platforms are transforming biomarker discovery.
Understanding Protein Biomarkers
Types of Protein Biomarkers
Researchers classify protein biomarkers into several categories based on their roles in disease research. Each type provides unique insights into biological processes and disease mechanisms. The table below summarizes the main categories and their descriptions:
| Category | Description |
| Susceptibility and Risk | Indicates the likelihood of developing a disease. |
| Diagnostic | Used to confirm the presence of a disease. |
| Monitoring | Tracks the progression of a disease or response to treatment. |
| Prognostic | Provides information about the likely outcome of a disease. |
| Predictive | Helps to predict the response to a specific treatment. |
| Pharmacodynamic and Treatment Response | Assesses the effects of a treatment on the biological system. |
| Safety | Evaluates the safety of a treatment or intervention. |
Diagnostic Biomarkers
Diagnostic biomarkers help researchers confirm the presence of a disease. They detect specific proteins that appear or change when a disease develops. Scientists use these markers to distinguish between healthy and diseased samples in research studies.
Prognostic Biomarkers
Prognostic biomarkers provide information about the likely outcome or course of a disease. These proteins can indicate whether a disease will progress quickly or slowly. Researchers use them to group samples based on expected disease behavior.
Predictive Biomarkers
Predictive biomarkers help determine how a biological system might respond to a specific treatment. They guide researchers in selecting the most promising experimental interventions. These markers support the design of targeted studies.
Pharmacodynamic Biomarkers
Pharmacodynamic biomarkers assess the effects of a treatment on biological systems. They show how proteins change in response to experimental drugs or interventions. Scientists use these markers to evaluate the impact of new compounds in preclinical research.
Biological Roles and Relevance in Disease
Protein biomarkers play key roles in many biological processes. Their presence or changes often reflect underlying disease mechanisms. The table below highlights some common biological processes associated with protein biomarkers:
| Biological Process | Description |
| Inflammation | Proteins involved in immune responses that indicate health deterioration after stroke. |
| Coagulation | Proteins that play a role in blood clotting, which can be disrupted in disease progression. |
| Immune Responses | Proteins that are part of the body's defense mechanism, indicating changes in disease states. |
Researchers often focus on proteins linked to inflammation, coagulation, and immune responses. These processes frequently change during disease development and progression. For example, inflammation-related proteins can signal early biological changes in cancer or other complex diseases.
Tip: High-throughput technologies such as olink proteomics multiplex biomarker panels allow scientists to measure thousands of protein biomarkers across these biological processes, supporting large-scale discovery and validation studies.
By understanding the types and roles of protein biomarkers, research teams can design more effective experiments and uncover new insights into disease biology.
Key Concepts in Protein Biomarker Detection
Sensitivity and Dynamic Range
Sensitivity describes how well a detection method identifies low-abundance proteins in a sample. Dynamic range refers to the span of concentrations that an assay can measure accurately. Researchers often encounter challenges when detecting proteins present at very low levels, especially in body fluids. Many diagnostic protein analytes exist in concentrations below the detectable limits of standard assays. Mass spectrometry, for example, struggles with poor sensitivity for low-abundance biomarkers. Affinity enrichment methods, such as using specific antibodies, can improve sensitivity, but these approaches have limitations.
The following table summarizes how sensitivity and dynamic range impact the detection of low-abundance protein biomarkers:
| Evidence Description | Impact on Detection |
| Mass spectrometry has poor sensitivity for low abundant biomarkers in body fluids. | High sensitivity is crucial for detecting low-abundance biomarkers. |
| Most diagnostic protein analytes exist in low concentrations, often below detectable limits. | Effective sensitivity needs improvement for routine clinical diagnostics. |
| Affinity enrichment methods enhance sensitivity but have drawbacks. | These methods are essential for improving detection of low-abundance biomarkers. |
| Ideal workflows should include enhanced chromatography and statistical analysis. | Increased sensitivity will facilitate the discovery of disease risk biomarkers. |
| Typical detection modalities have limited quantitative signal resolution. | This constrains the ability to detect low-abundance analytes effectively. |
| The signal from lower-abundance analytes is often masked by background noise. | This results in little to no detectable signal for low-abundance biomarkers. |
| Affinity reagents have binding curves modeled by the Langmuir isotherm. | This affects the dynamic range of detection methods. |
| The quantitative signal resolution is limited to 3–4 orders of magnitude. | This constrains the output signal curve achievable for low-abundance biomarkers. |
Researchers should select platforms with high sensitivity and broad dynamic range to maximize the detection of disease-associated proteins. Technologies such as olink proteomics multiplex biomarker panels offer improved sensitivity and dynamic range, enabling the quantification of both high- and low-abundance proteins in a single assay.
Specificity and Cross-Reactivity
Specificity measures how accurately an assay detects the intended protein target without interference from similar molecules. Cross-reactivity occurs when an antibody or detection reagent binds to unintended proteins, which can lead to false positives and reduced assay accuracy. Polyclonal antibodies often cause cross-reactivity because they recognize multiple epitopes. This problem highlights the importance of careful antibody selection in assay design.
Researchers can improve specificity by using monoclonal antibodies. These reagents target a single epitope, which reduces the risk of cross-reactivity and enhances the reliability of assay results. High specificity is essential for accurate protein quantification, especially in complex biological samples.
Tip: Careful validation of antibody reagents and assay protocols helps minimize cross-reactivity and ensures robust data quality.
Multiplexing Capability
Multiplexing capability refers to the number of proteins that a platform can measure simultaneously. High multiplexing allows researchers to analyze many biomarkers in a single experiment, saving time and sample volume. Commercial platforms vary in their multiplexing performance, sensitivity, and sample requirements.
The table below compares the multiplexing capability of several leading platforms:
| Platform | Sensitivity | Sample Volume | Correlation Coefficient |
| NULISA | Attomolar | 10 µL | 0.71 to 0.97 |
| MSD | Moderate | Varies | Moderate to Strong |
| Olink | High | Varies | Moderate to Strong |
- Pairwise Pearson correlation and intraclass correlation coefficient (ICC) analyses show good comparability across platforms.
- Proteins such as CXCL8, IL18, VEGFA, and MMP-1 demonstrate strong agreement, with correlation coefficients ranging from 0.71 to 0.97.
- Most platforms exhibit moderate to strong correlations for detected proteins, indicating consistency in protein expression measurements.
Multiplexing capability is a key advantage for large-scale studies. Researchers can profile hundreds or thousands of proteins, which accelerates biomarker discovery and validation. High-throughput platforms support efficient workflows and enable comprehensive analysis of complex biological samples.
Sample Volume and Matrix Considerations
Sample volume and matrix type play a critical role in protein biomarker analysis. Researchers must consider these factors when designing experiments for multiplex biomarker panels or high-throughput proteomics platforms like Olink. The amount of sample available often limits the number of proteins that can be measured, especially in studies using precious biological materials such as plasma, serum, or tissue lysates.
Small sample volumes require highly sensitive detection methods. Many advanced platforms, including Olink, can quantify thousands of proteins from just a few microliters of sample. This capability allows researchers to conserve valuable specimens while still obtaining comprehensive protein profiles. When working with limited sample amounts, scientists must select assays with a broad dynamic range and high reproducibility.
Matrix type refers to the biological environment from which the sample is derived. Common matrices include plasma, serum, cerebrospinal fluid, urine, and tissue extracts. Each matrix contains unique proteins, salts, and other molecules that can influence assay performance. For example, plasma contains high levels of albumin and immunoglobulins, which may interfere with the detection of low-abundance biomarkers. Researchers must optimize sample preparation protocols to minimize matrix effects and improve quantification accuracy.
The following table summarizes how sample volume and matrix type influence protein biomarker assay performance:
| Aspect | Findings |
| Sample Volume Impact | Quantification accuracy was higher in data-independent acquisition (DIA) analyses compared to data-dependent acquisition (DDA), especially with low protein amounts. |
| Matrix Type Influence | Smaller spectral libraries improved quantification specificity, while larger libraries reduced it. |
| Reproducibility Comparison | DIA showed better reproducibility than DDA due to lower interference and the stochastic nature of DDA. |
| Accuracy Discrepancy | Differences in accuracy were noted between peptide and protein levels, indicating challenges in protein inference. |
Researchers often choose data-independent acquisition (DIA) workflows for studies with small sample volumes. DIA provides higher quantification accuracy and reproducibility than traditional data-dependent acquisition (DDA) methods. Smaller spectral libraries can also improve specificity, which is important when analyzing complex matrices.
Sample matrix selection affects not only assay sensitivity but also data interpretation. For example, urine and cerebrospinal fluid may contain fewer interfering proteins than plasma, making them suitable for certain biomarker discovery projects. However, each matrix presents unique challenges, such as variable protein concentrations and the presence of endogenous inhibitors.
Tip: Scientists should validate their protein biomarker assays in the specific matrix of interest to ensure reliable results. Careful optimization of sample preparation and assay conditions can reduce matrix effects and improve reproducibility.
Common Methods for Protein Biomarker Detection
Immunoassay-Based Methods
ELISA
Enzyme-linked immunosorbent assay (ELISA) remains a foundational technique for protein biomarker detection. Researchers use ELISA to measure specific proteins in biological samples. The method relies on antibodies that bind to the target protein, followed by an enzyme reaction that produces a measurable signal. ELISA offers high sensitivity and specificity, making it suitable for quantifying low-abundance proteins. However, traditional ELISA assays typically analyze one protein per experiment, which limits throughput in large-scale studies.
Multiplex Immunoassays (e.g., Luminex, MSD)
Multiplex immunoassays, such as Luminex and Meso Scale Discovery (MSD), allow simultaneous measurement of multiple proteins in a single sample. These platforms use color-coded beads or patterned arrays, each coupled with a specific antibody. Researchers can profile dozens to hundreds of biomarkers at once, conserving sample volume and increasing efficiency. Multiplex immunoassays provide moderate to high sensitivity and are widely used for screening panels in cancer research and other disease studies.
Advantages and Limitations
Immunoassay-based methods offer several advantages:
- High sensitivity for detecting low-abundance proteins.
- Strong specificity due to antibody-antigen interactions.
- Established protocols and broad availability.
Limitations include:
- Single-plex ELISA restricts throughput.
- Multiplex assays may experience cross-reactivity between antibodies.
- Assay performance depends on antibody quality and sample matrix.
Mass Spectrometry–Based Detection
Discovery Proteomics (LC–MS/MS)
Liquid chromatography-tandem mass spectrometry (LC–MS/MS) enables researchers to identify and quantify thousands of proteins in complex samples. Discovery proteomics uses unbiased workflows to detect novel biomarkers and characterize protein expression patterns. This approach excels at profiling post-translational modifications and protein isoforms, providing deep insights into biological processes.
Targeted Proteomics (MRM/SRM)
Multiple reaction monitoring (MRM) and selected reaction monitoring (SRM) are targeted mass spectrometry techniques. Researchers use these methods to quantify specific proteins with high precision. Targeted proteomics offers robust reproducibility and is ideal for validating candidate biomarkers identified in discovery studies.
Advantages and Limitations
Researchers compare mass spectrometry and immunoassays using sensitivity and specificity metrics:
| Technique | Sensitivity | Specificity |
| Mass Spectrometry-based Immunoassays (MSIA) | Lower sensitivity compared to sandwich immunoassays | High specificity, can identify protein sequences and structures, including post-translational modifications |
| Sandwich Immunoassays (e.g., ELISA) | Higher sensitivity, capable of detecting proteins at lower concentrations | High specificity, but lower throughput as one protein is assayed at a time |
Mass spectrometry provides high specificity and can characterize protein structures and modifications. However, it often shows lower sensitivity for low-abundance proteins compared to immunoassays. Researchers select the appropriate method based on study goals, sample type, and required throughput.
High-Plex Affinity-Based Protein Assays
Proximity-Based Technologies
High-plex affinity-based assays use proximity-based technologies to improve specificity and multiplexing. Olink, for example, employs proximity extension assays that require two antibodies to bind the target protein. This dual recognition reduces cross-reactivity and enables high-throughput measurement of thousands of proteins.
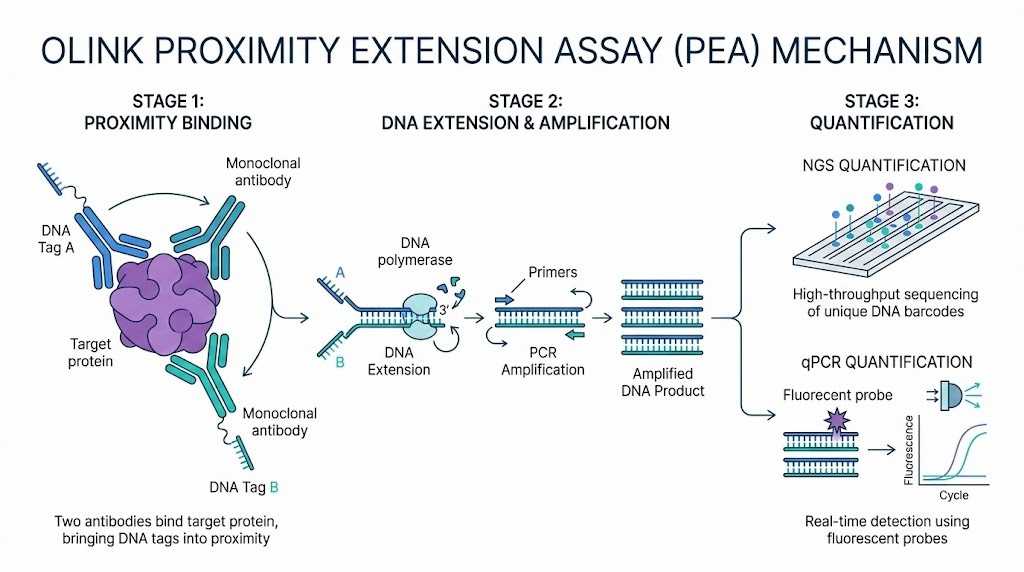
Ultra-Sensitive Low-Volume Platforms
Recent advances have produced platforms that deliver ultra-sensitive detection from minimal sample volumes. These systems, such as olink proteomics multiplex biomarker panels, allow researchers to profile over 5,000 proteins using just a few microliters of sample. This capability is essential for studies with limited biological material.
Advantages for Biomarker Studies
Researchers benefit from high-plex affinity-based assays in several ways:
- They achieve broad proteome coverage and robust quantification.
- They minimize matrix effects and improve reproducibility.
- They support multi-omics integration for comprehensive analysis.
The following table summarizes key features and performance metrics of leading high-plex assays:
| Assay Type | Key Features | Performance Metrics |
| SomaLogic | High-affinity aptamer binders for each target protein | Variable specificity depending on binding epitope and sample matrix |
| Olink | Proximity extension assays requiring two antibodies for specificity | Mitigates specificity issues, allowing for high-throughput measurements |
| NULISA | Higher sensitivity and lower limit of detection compared to other methods | Not as extensive in proteome coverage but excels in sensitivity |
| MS-based Proteomics | Less matrix-sensitive, capable of identifying PTMs and protein isoforms | Limited depth for plasma analysis but improved proteome coverage and throughput with recent advancements |
| Seer Proteograph™ | Uses surface-modified magnetic nanoparticles for protein enrichment | Significantly increases soluble proteome coverage |
| nELISA | High-throughput, cost-effective, and scalable for multimodal screening | Offers robust quantification of secreted proteins, enhancing drug discovery efforts |
Researchers select high-plex platforms to accelerate biomarker discovery and validation, especially in cancer screening and large-scale population studies.
Other Detection Approaches
Western Blot
Western blotting remains a classic method for protein biomarker analysis. Researchers use this technique to separate proteins by size through gel electrophoresis. After separation, they transfer proteins to a membrane and probe with specific antibodies. This approach allows for the detection and semi-quantitative analysis of target proteins. Western blotting provides high specificity when using well-validated antibodies. Scientists often use this method to confirm the presence of a protein identified by other assays. However, Western blotting has limited throughput and requires relatively large amounts of sample. It is best suited for targeted studies rather than large-scale biomarker discovery.
Immunohistochemistry (IHC)
Immunohistochemistry (IHC) enables researchers to visualize protein expression within tissue sections. This method uses antibodies tagged with enzymes or fluorescent markers to bind specific proteins in situ. IHC reveals the spatial distribution of biomarkers, which is valuable for understanding disease mechanisms at the cellular level. Scientists can compare protein expression in healthy and diseased tissues, gaining insights into cancer progression and tissue architecture. IHC supports multiplexing by using multiple antibodies with different labels, but the number of targets analyzed in a single experiment remains limited. Researchers value IHC for its ability to link protein biomarkers to histological context.
Biosensor-Based Methods
Biosensor-based methods are emerging as powerful tools for protein biomarker detection. These platforms use physical or chemical sensors to detect biomolecular interactions in real time. Surface plasmon resonance (SPR), electrochemical sensors, and microfluidic chips are common examples. Biosensors offer label-free detection and can measure binding events with high sensitivity. Recent advances have enabled the detection of proteins at femtogram levels, expanding the range of measurable biomarkers.
Researchers have developed blood-based biomarker detection methods that provide non-invasive options for sample collection. For example, ultrasensitive biosensors can detect neuronal proteins such as neurofilament light chain (NfL) in blood, which supports studies of disease activity in neurological research. Saliva has also emerged as a promising biofluid for protein biomarker analysis. Scientists have identified markers like total tau (t-tau) and glial fibrillary acidic protein (GFAP) in saliva, which may reflect disease processes in neurodegeneration.
Key advantages of biosensor-based approaches include:
- Real-time, label-free detection of protein interactions
- Compatibility with small sample volumes and diverse biofluids
- Potential for multiplexing and integration with microfluidic systems
Advances in immunoassays and biosensor technologies continue to enhance the sensitivity and multiplexing capability of protein biomarker analysis. These methods support the discovery of novel biomarkers in cancer screening and other research fields.
| Method | Key Feature | Typical Application |
| Western Blot | Protein separation and detection | Target validation |
| Immunohistochemistry | Spatial localization in tissues | Tissue-based biomarker studies |
| Biosensor-Based | Real-time, label-free measurement | High-sensitivity biomarker assays |
Note: Researchers should select the detection approach that best matches their experimental goals, sample type, and desired throughput.
Comparison of Protein Biomarker Detection Methods
Comparison by Sensitivity
Sensitivity determines how well a detection method identifies proteins present at very low concentrations. Researchers often select assays with high sensitivity to detect early changes in disease states. ELISA stands out as a gold standard, achieving detection limits as low as 1 picogram per milliliter. This high sensitivity enables the quantification of low-abundance biomarkers, which is essential for cancer screening and discovery. Multiplexed platforms also perform well for proteins present in higher concentrations, but their sensitivity can decrease for targets like IL4, IL10, and IL13. The following table summarizes the analytical sensitivity of common methods:
| Method | Detection Limit (DL) | Strengths | Limitations |
| ELISA | Approaches 1 pg mL−1 | High sensitivity and specificity | Requires technical expertise, limited point-of-care application |
| Multiplexed Platforms | Varies (high for abundant proteins) | Efficient for multiple targets | Lower sensitivity for some low-abundance proteins |
Researchers must consider sensitivity benchmarks when designing experiments for protein biomarker analysis, especially in studies focused on early cancer detection.
Comparison by Specificity
Specificity refers to the ability of a method to distinguish the target protein from similar molecules. High specificity reduces false positives and improves data reliability. ELISA and fluorescent bead immunoassays both offer strong specificity due to antibody-antigen interactions. Mass spectrometry provides excellent specificity by identifying unique peptide sequences, making it valuable for confirming protein identity. Microarray immunoassays deliver moderate specificity, which may require additional validation steps. Researchers often select methods with high specificity for studies involving complex biological samples, such as plasma or serum.
| Technology Type | Specificity | Notes |
| Mass Spectrometry | High | Identifies unique peptides, suitable for validation |
| Fluorescent Bead Immunoassays | High | Strong antibody-antigen binding |
| ELISA | High | Gold standard for specificity |
| Microarray Immunoassays | Moderate | May require further validation |
Comparison by Multiplexing Capability
Multiplexing capability describes how many proteins a platform can measure in a single experiment. Researchers value high multiplexing for large-scale studies and comprehensive biomarker panels. Microarray immunoassays and fluorescent bead platforms allow simultaneous analysis of dozens to hundreds of proteins, reducing sample volume and increasing throughput. Mass spectrometry supports moderate multiplexing, excelling in candidate identification but offering lower throughput for clinical applications. ELISA traditionally measures one analyte per assay, but newer formats enable limited multiplexing.
| Technology Type | Multiplexing Capability | Throughput | Notes |
| Mass Spectrometry | Moderate | Low | Best for candidate identification, limited clinical throughput |
| Fluorescent Bead Immunoassays | High | High | Suitable for panels with multiple analytes |
| ELISA | Low to Moderate | Moderate | Single analyte standard, some multiplexing possible |
| Microarray Immunoassays | Very High | Very High | Analyzes many targets with reduced sample volume |
Bar chart illustrating multiplexing throughput among leading protein biomarker detection technologies. Source: PubMed/NCBI scientific literature.
Tip: High multiplexing platforms, such as microarray immunoassays and fluorescent bead systems, support efficient workflows for protein biomarker analysis in cancer screening and discovery.
Comparison by Sample Requirements
Protein biomarker detection methods differ in the amount and type of sample they require. ELISA and traditional immunoassays often need larger sample volumes, sometimes up to 100 microliters per target. This requirement can limit their use in studies with scarce or precious samples, such as pediatric or rare disease cohorts. Multiplex immunoassays, including bead-based and planar array platforms, reduce sample consumption by measuring multiple proteins in a single reaction. These systems typically require 10–50 microliters per assay.
High-plex affinity-based platforms, such as Olink's Proximity Extension Assay (PEA), excel in minimizing sample requirements. Researchers can analyze thousands of proteins using only a few microliters of plasma, serum, or other biofluids. This efficiency supports large-scale studies and enables repeated measurements from limited sample sources. Mass spectrometry-based methods show flexibility in sample input but may need higher protein concentrations for optimal sensitivity, especially in discovery workflows.
Sample matrix also influences method selection. Plasma, serum, urine, and tissue lysates each present unique challenges. High-abundance proteins in plasma can mask low-abundance biomarkers, requiring careful sample preparation. Some platforms, like PEA, demonstrate robust performance across diverse matrices, supporting broad research applications. Researchers should validate assay performance in the intended matrix to ensure accurate quantification.
Comparison by Throughput
Throughput describes how many samples or analytes a method can process in a given time. High-throughput capability is essential for large-scale cancer screening and biomarker discovery projects. Researchers often select platforms based on their ability to handle hundreds or thousands of samples efficiently.
- DSP Technology enables high-plex spatial profiling in formalin-fixed, paraffin-embedded (FFPE) tissue samples. This method quantifies proteins in different tissue compartments without destroying the sample.
- Proximity Extension Assay (PEA), as used in Olink's Protein Biomarker Profiling Service, offers high-throughput detection in liquid biopsies. PEA uses DNA-encoded tags for specificity and sensitivity, covering a broad dynamic range with minimal sample input.
Multiplex immunoassays and microarray platforms also support high-throughput workflows, allowing simultaneous measurement of many proteins across large cohorts. Mass spectrometry, while powerful for discovery, may require longer run times and more complex data analysis, which can limit throughput in some settings.
Tip: High-throughput platforms accelerate biomarker discovery by enabling rapid, large-scale protein profiling. This advantage is critical for population studies and longitudinal research.
Comparison by Suitability for Cancer Screening
Suitability for cancer screening depends on several factors, including sensitivity, specificity, throughput, and the ability to detect early disease changes. Researchers prioritize methods that identify low-abundance biomarkers and support multiplex analysis. High-plex affinity-based assays, such as Olink's PEA, provide strong performance in these areas. These platforms detect subtle protein changes in early-stage cancer and enable comprehensive panel development.
Studies show that clinical utility often aligns with clinical validity when a new test identifies a similar disease spectrum as established standards. However, direct evidence of reduced mortality or morbidity remains essential, especially when new technologies enter the field. The evolution of cervical cancer screening programs illustrates how accurate, validated tests—like Pap smears and HPV assays—can reduce disease incidence when integrated into well-designed research and public health strategies.
Researchers should select detection methods that combine high sensitivity, specificity, and throughput. These features ensure robust biomarker discovery and support the development of effective cancer screening tools.
Summary Table: Advantages and Limitations of Each Method
Researchers often compare protein biomarker detection methods using several key criteria. These include sensitivity, specificity, multiplexing capability, sample volume requirements, and suitability for different research applications. The following tables summarize the main advantages and limitations of each method, providing a quick reference for selecting the optimal approach in protein biomarker analysis.
Sensitivity vs. Specificity Table
| Method | Sensitivity | Specificity | Key Notes |
| ELISA | High (pg/mL) | High | Gold standard for single-analyte detection; robust for low-abundance proteins |
| Multiplex Immunoassays | Moderate to High | Moderate to High | Enables simultaneous detection; may have cross-reactivity in complex samples |
| Mass Spectrometry (LC–MS/MS) | Moderate | Very High | Excellent for protein identification and PTMs; less sensitive for very low-abundance targets |
| High-Plex Affinity-Based (Olink, SomaLogic) | Very High (sub-pg/mL) | Very High | Combines high sensitivity with specificity; ideal for large-scale biomarker panels |
| Biosensor-Based Methods | High | High | Real-time, label-free detection; suitable for rapid screening |
Note: High-plex affinity-based platforms such as Olink offer both high sensitivity and specificity, making them valuable for multiplex biomarker panels in cancer research.
References: PubMed/NCBI scientific literature.
Multiplexing vs. Sample Volume Table
| Method | Multiplexing Capability | Sample Volume Required | Research Value |
| ELISA | Low (1–2 analytes) | Moderate to High (50–100 µL) | Best for targeted validation |
| Multiplex Immunoassays | Moderate (10–100 analytes) | Low to Moderate (10–50 µL) | Efficient for mid-scale panels |
| Mass Spectrometry | High (100s–1000s analytes) | Moderate (10–100 µg protein) | Discovery proteomics; broad coverage |
| High-Plex Affinity-Based (Olink) | Very High (up to 5400 analytes) | Very Low (1–3 µL) | Ideal for large-scale studies with limited samples |
| Biosensor-Based Methods | Moderate | Low | Suitable for rapid, small-scale screening |
Tip: Olink's high-plex technology enables comprehensive protein biomarker analysis from minimal sample volumes, supporting studies with scarce biological material.
Application Suitability Table
| Method | Discovery Proteomics | Targeted Validation | High-Throughput Screening | Cancer Screening & Discovery |
| ELISA | Limited | Excellent | Limited | Moderate |
| Multiplex Immunoassays | Moderate | Good | Good | Good |
| Mass Spectrometry | Excellent | Good | Moderate | Good |
| High-Plex Affinity-Based (Olink) | Excellent | Excellent | Excellent | Excellent |
| Biosensor-Based Methods | Moderate | Moderate | Good | Moderate |
Researchers should match the detection method to their experimental goals. High-plex affinity-based assays, such as Olink, provide unmatched versatility for both discovery and validation phases in protein biomarker analysis. These platforms support multiplex biomarker panels, high-throughput workflows, and integration with multi-omics strategies, which are essential for advancing cancer screening and discovery.
For further details on technical performance and workflow optimization, consult recent reviews in PubMed/NCBI scientific literature.
Protein Biomarker Analysis in Cancer Screening
Role of Protein Biomarkers in Early Cancer Detection
Protein biomarkers play a central role in the early detection of cancer. Researchers use these molecules to identify disease before symptoms appear. Early detection increases the likelihood of successful intervention and improves research outcomes. Scientists have identified several protein biomarkers that serve different purposes in cancer studies. The table below summarizes their main roles, examples, and significance:
| Role of Protein Biomarkers | Examples | Significance |
| Early Detection | PSA, CA-125, AFP | Enables prompt research intervention and increases chances of effective study |
| Monitoring Treatment | HER2, PSA, CA-125 | Assesses response to experimental therapies and guides research adjustments |
| Assessing Prognosis | HER2, PSA, CA-125 | Provides insights into disease stage and aggressiveness for research analysis |
Researchers often select panels of protein biomarkers to improve the accuracy of early cancer detection. These panels help identify subtle changes in protein expression that may signal the onset of disease. By tracking these changes, scientists can design better experiments and develop new strategies for cancer screening.
Challenges in Cancer Biomarker Detection
Cancer biomarker detection presents several significant challenges for research teams:
- Low sensitivity and specificity of tests, especially in early-stage cancers.
- Variability in test results across different platforms and laboratories.
- Ethnic disparities in biomarker expression.
- Biomarkers may vary between patients or within different regions of the same tumor.
- Inconsistent results can skew research findings or experimental decisions.
- Rigorous validation and qualification are required for regulatory approval.
- Challenges in securing reimbursement from funding agencies.
- Biomarkers must be validated in diverse populations.
- Standardization of testing protocols and laboratory practices is essential.
- Measurement of protein biomarkers is limited by detection methods like ELISA.
- Clinical translation is often stalled by the lack of well-characterized monoclonal antibodies.
These challenges highlight the need for robust, reproducible, and sensitive detection methods. Researchers must address variability and standardization to ensure reliable results across studies.
Importance of High-Sensitivity and Multiplex Panels
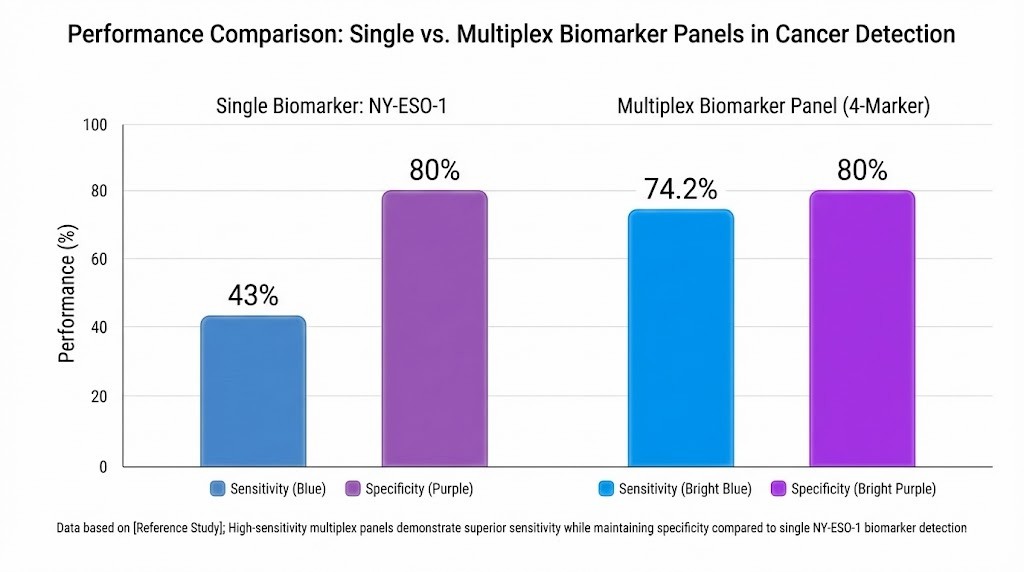
High-sensitivity and multiplex panels have transformed cancer biomarker research. Traditional single-analyte assays often miss early-stage cancers due to limited sensitivity. Multiplex panels allow researchers to measure several biomarkers at once, increasing the likelihood of detecting cancer at an early stage. The table below compares the performance of single-analyte and multiplex assays:
| Assay Type | Sensitivity | Specificity | Early Stage Sensitivity | Late Stage Sensitivity |
| NY-ESO-1 (single) | 43% | 80% | 44.7% | 34.5% |
| 4-biomarker panel (NY-ESO-1 + 3 markers) | 74.2% | 80% | 72.7% | 82.8% |
*Grouped bar chart comparing sensitivity and specificity of single-analyte and multiplex cancer detection assays. Source: PubMed/NCBI scientific literature.*
Multiplex panels, such as those enabled by olink proteomics multiplex biomarker panels, provide higher sensitivity for both early and late-stage cancers. These platforms support the simultaneous analysis of thousands of proteins, which helps researchers identify complex biomarker signatures. High-sensitivity detection ensures that even low-abundance proteins, which may serve as early indicators of disease, are not overlooked. This approach accelerates biomarker discovery and improves the reliability of cancer screening studies.
Technologies Best Suited for Cancer Screening
Researchers have identified several advanced technologies that excel in protein biomarker analysis for cancer screening. These platforms combine high sensitivity, specificity, and throughput, making them valuable for large-scale studies and early detection research.
Key technologies include:
- High-Plex Affinity-Based Protein Assays:
Platforms such as Olink's Protein Biomarker Profiling Service use proximity extension assays to measure thousands of proteins in a single run. This technology enables researchers to analyze over 5,400 proteins from minimal sample volumes. The broad dynamic range and robust reproducibility support the detection of both high- and low-abundance biomarkers, which is essential for early cancer research. - Mass Spectrometry–Based Proteomics:
Liquid chromatography-tandem mass spectrometry (LC–MS/MS) and targeted proteomics approaches allow for comprehensive protein profiling. These methods provide deep coverage of the proteome and can identify post-translational modifications. While mass spectrometry offers high specificity, its sensitivity for low-abundance proteins may be lower than affinity-based assays. - Multiplex Immunoassays:
Technologies such as Luminex and MSD enable simultaneous quantification of multiple protein biomarkers. These platforms are suitable for mid-scale panels and offer moderate to high sensitivity. Researchers often use them for validation studies and targeted screening. - Artificial Intelligence (AI)–Supported Platforms:
AI technologies have shown strong performance in cancer screening, especially in imaging-based applications like mammography. In recent studies, AI-supported mammography achieved a breast cancer detection rate of 6.7 per 1,000, which is 17.6% higher than the control group. The recall rate was lower in the AI group (37.4 per 1,000) compared to controls (38.3 per 1,000). The positive predictive value (PPV) of recall reached 17.9% in the AI group, surpassing the control group's 14.9%. The PPV of biopsy was also higher at 64.5% versus 59.2%. These metrics highlight the potential of AI to enhance accuracy and efficiency in cancer screening workflows.
Note: Researchers should select technologies based on study goals, sample availability, and the need for multiplex biomarker panels. High-throughput proteomics platforms like Olink offer unmatched scalability and integration with multi-omics strategies, supporting discovery and validation in cancer research.
| Technology Type | Multiplexing Capability | Sensitivity | Throughput | Research Value |
| High-Plex Affinity-Based (Olink) | Very High (up to 5,400) | Very High | High | Early detection, large-scale biomarker panels |
| Mass Spectrometry (LC–MS/MS) | High | Moderate to High | Moderate | Proteome coverage, PTM analysis |
| Multiplex Immunoassays (Luminex, MSD) | Moderate (10–100) | Moderate to High | High | Targeted panels, validation studies |
| AI-Supported Imaging | N/A (Imaging-based) | High (for imaging) | High | Enhanced detection rates, workflow efficiency |
Researchers continue to advance protein biomarker analysis by integrating high-throughput proteomics, multiplex biomarker panels, and AI-driven tools. These technologies drive progress in cancer screening and discovery, enabling more comprehensive and reliable research outcomes.
Protein Biomarker Discovery Strategies
Discovery Proteomics Workflow
Researchers use a systematic workflow to identify and advance protein biomarker candidates. This process begins with non-targeted proteomics, which enables the discovery of a large pool of potential biomarkers. Scientists then confirm statistically significant differences in protein abundance between disease and control groups. They validate selected targets using methods such as parallel reaction monitoring (PRM) and enzyme-linked immunosorbent assay (ELISA). The workflow continues with assay optimization for research applications and concludes with clinical validation and commercialization for research tools.
| Phase | Description |
| Discovery | Identify a large pool of candidate biomarkers using non-targeted proteomics. |
| Qualification | Confirm statistically significant abundance differences between disease and control groups. |
| Verification | Validate selected target proteins using various methods such as MS-based PRM and ELISA. |
| Research Assay Optimization | Optimize assays for research purposes. |
| Clinical Validation | Confirm the practical utility of the biomarker assay in research settings. |
| Commercialization | Move towards the commercialization of validated biomarkers for research use. |
This structured approach ensures that each phase builds on the previous one, increasing the reliability of protein biomarker analysis in cancer screening and discovery.
Validation Strategies for Candidate Biomarkers
Validation of candidate biomarkers requires robust statistical and analytical methods. Scientists often use logistic regression to assess the relationship between biomarker levels and disease status. Principal component analysis (PCA) helps identify patterns in biomarker expression, explaining a large portion of variance in the data. Targeted mass spectrometry, such as PRM-MS, enables researchers to confirm the predictive value of biomarkers in multivariate models.
| Validation Strategy | Description |
| Logistic Regression | Assesses the relationship between candidate biomarkers and disease diagnosis. |
| Principal Component Analysis (PCA) | Identifies primary components explaining variance in biomarker expression. |
| Targeted Mass Spectrometry (PRM-MS) | Validates biomarkers through predictive models, assessing their ability to detect disease. |
These strategies provide a rigorous foundation for advancing multiplex biomarker panels and high-throughput proteomics platforms in research.
Integrating High-Plex Protein Detection Platforms
High-plex protein detection platforms play a critical role in modern biomarker discovery pipelines. Researchers integrate technologies such as Olink's Protein Biomarker Profiling Service to systematically credential biomarker candidates. This integration involves several key steps:
- Pre-processing data to remove noise and ensure data integrity.
- Applying proprietary normalization algorithms to adjust for technical variation.
- Using statistical tools for hypothesis testing and significance determination.
- Employing ROC analysis to evaluate biomarker performance and select optimal thresholds.
- Implementing regression models and decision trees to translate multiplex biomarker signals into actionable research results.
Scientists often begin with high-performance liquid chromatography-tandem mass spectrometry (LC-MS/MS) for candidate discovery. They then qualify biomarkers analytically and verify them quantitatively using methods like SID-MRM-MS. The pipeline transitions from proximal fluid analysis to peripheral blood, enabling verification in diverse sample types.
This systematic approach enhances the robustness of protein biomarker analysis, supports the development of multiplex biomarker panels, and accelerates high-throughput proteomics research in cancer screening and discovery.
Choosing the Right Protein Biomarker Detection Method
Matching Methods to Research Goals
Selecting the optimal protein biomarker detection method depends on the specific objectives of a research project. Scientists often distinguish between discovery and targeted validation studies. In discovery workflows, researchers seek to identify novel protein biomarkers across large sample sets. High-throughput proteomics platforms, such as Olink's Protein Biomarker Profiling Service, enable broad screening of thousands of proteins. These platforms support multiplex biomarker panels and facilitate the identification of early biological changes in cancer research.
Discovery vs. Targeted Validation
Discovery studies require methods that offer extensive proteome coverage and unbiased detection. Mass spectrometry-based approaches and high-plex affinity-based assays excel in this context. Researchers use these technologies to generate comprehensive protein profiles and uncover new biomarker candidates. In contrast, targeted validation studies focus on confirming the relevance of selected proteins. Scientists often employ ELISA or multiplex immunoassays for precise quantification of specific biomarkers. These methods provide robust sensitivity and specificity, supporting the verification of candidate proteins identified during discovery.
Clinical Research vs. Mechanistic Studies
Research teams also consider whether their goals align with clinical translation or mechanistic investigation. Clinical research projects prioritize reproducibility, scalability, and regulatory compliance. High-plex platforms, such as Olink, offer standardized workflows and high data quality, which are essential for large-scale studies. Mechanistic studies, on the other hand, may require detailed analysis of protein interactions, post-translational modifications, or pathway dynamics. Mass spectrometry and biosensor-based methods provide valuable insights into protein function and regulation.
Decision Framework for Selecting Detection Technologies
Researchers use structured decision frameworks to guide the selection of protein biomarker detection technologies. Feature selection methods play a critical role in translating discoveries from untargeted mass spectrometry into clinically relevant panels. These approaches help scientists narrow down large datasets to a manageable number of proteins for further validation. Decision trees offer a systematic way to evaluate biomarker candidates and their affinity reagents. By applying these frameworks, research teams can identify the most promising proteins for assay development and ensure that selected methods align with project requirements.
| Decision Framework | Application | Benefit |
| Feature Selection | Clinical translation of biomarkers | Reduces complexity, focuses validation |
| Decision Trees | Candidate selection and validation | Systematic investigation, assay design |
Tip: A well-defined decision framework improves the efficiency and reliability of protein biomarker analysis, supporting the transition from discovery to validation.
Why High-Plex, High-Sensitivity Protein Assays Are Often Preferred
High-plex, high-sensitivity protein assays have become the preferred choice for many research professionals. These platforms offer several advantages:
- Accurate quantification of proteins, which is essential for understanding complex biological systems.
- Direct observation of protein dynamics, addressing limitations of genomics and transcriptomics.
- Enhanced specificity in multiplex formats, ensuring reliable measurement of intended analytes.
- Assessment and mitigation of cross-reactivity, which maintains assay integrity.
- Efficient use of time and resources, balancing assay development with the benefits of multiplexing.
Researchers recognize that protein profiling methods provide a more direct view of biological processes than nucleic acid-based approaches. High-plex assays, such as those offered by Olink, enable comprehensive analysis from minimal sample volumes. This capability supports large-scale studies and accelerates biomarker discovery in cancer screening and other fields.
Note: The preference for high-plex, high-sensitivity assays reflects the need for scalable, reproducible, and data-rich workflows in modern protein biomarker analysis. Scientists should match detection technologies to their research goals, sample availability, and desired throughput to maximize the impact of their studies.
Limitations of Traditional Biomarker Assays
Single-Plex Limitations (ELISA)
Researchers have relied on enzyme-linked immunosorbent assay (ELISA) for decades to quantify protein biomarkers. ELISA provides high sensitivity and specificity for single analytes. However, this method restricts throughput because each assay measures only one protein at a time. Large-scale studies in cancer screening and discovery often require analysis of hundreds or thousands of proteins. Single-plex ELISA workflows demand significant sample volume and labor, which limits efficiency. Scientists must run multiple assays in parallel to build multiplex biomarker panels, increasing cost and complexity. ELISA also struggles to detect low-abundance proteins when sample volume is limited. These constraints make single-plex ELISA less suitable for high-throughput proteomics and comprehensive protein biomarker analysis.
Note: Multiplex biomarker panels and high-throughput platforms address these limitations by enabling simultaneous measurement of many proteins from minimal sample input.
Cross-Reactivity in Multiplex Immunoassays
Multiplex immunoassays, such as bead-based or planar array systems, allow researchers to measure multiple proteins in one experiment. These platforms improve efficiency and conserve sample volume. However, cross-reactivity remains a significant challenge. Antibodies may bind to unintended targets, causing false positives or inaccurate quantification. Polyclonal antibodies, which recognize multiple epitopes, increase the risk of cross-reactivity. Even monoclonal antibodies can show off-target binding in complex biological matrices. Scientists must validate antibody specificity and optimize assay conditions to minimize interference.
Common sources of cross-reactivity:
- Shared epitopes among protein families
- High-abundance proteins masking low-abundance targets
- Matrix effects from plasma, serum, or tissue lysates
| Issue | Impact on Research |
| Cross-reactivity | Reduces assay accuracy |
| Matrix interference | Complicates data interpretation |
| Antibody variability | Limits reproducibility |
Multiplex immunoassays require rigorous validation to ensure reliable protein biomarker analysis, especially in cancer screening studies.
Constraints of Mass Spectrometry for Low-Abundance Targets
Mass spectrometry (MS) has revolutionized discovery proteomics by enabling unbiased identification of thousands of proteins. Researchers use MS to profile protein expression and post-translational modifications. Despite its strengths, MS faces challenges in detecting low-abundance biomarkers, particularly in plasma or serum. High-abundance proteins can suppress signals from rare targets, reducing sensitivity. Sample preparation and enrichment strategies help, but they add complexity and may not fully resolve the issue. Data-dependent acquisition (DDA) workflows often miss low-level proteins due to stochastic sampling. Data-independent acquisition (DIA) improves reproducibility, but quantification accuracy still depends on spectral library quality and sample complexity.
Tip: Scientists seeking comprehensive protein biomarker analysis in cancer research should consider platforms that combine high sensitivity, broad dynamic range, and robust multiplexing capability.
Multiplex biomarker panels and high-throughput proteomics technologies, such as proximity extension assays, offer scalable solutions for overcoming the limitations of traditional assays.
Need for Scalable High-Sensitivity Solutions
Modern protein biomarker analysis faces increasing demands for both scalability and sensitivity. Research teams often need to analyze thousands of proteins across hundreds or thousands of samples. Traditional single-plex assays, such as ELISA, cannot meet these requirements due to limited throughput and high sample consumption. Multiplex immunoassays and mass spectrometry offer improvements, but they still struggle with sensitivity for low-abundance proteins and scalability for large studies.
High-throughput proteomics platforms now address these challenges. Technologies like Olink's Protein Biomarker Profiling Service provide researchers with the ability to measure over 5,400 proteins in a single run. This approach uses proximity extension assays, which combine the specificity of immunoassays with the sensitivity of DNA amplification. As a result, scientists can detect both high- and low-abundance proteins from minimal sample volumes.
Key features of scalable high-sensitivity solutions include:
- Broad dynamic range: Researchers can quantify proteins that differ in concentration by up to ten orders of magnitude.
- Minimal sample input: Studies can proceed with as little as 1–3 microliters of plasma or serum.
- High reproducibility: Standardized workflows ensure consistent results across large cohorts.
- Multiplex biomarker panels: Scientists can analyze complex biological pathways and disease signatures in a single experiment.
Tip: Scalable, high-sensitivity platforms support multi-omics integration, allowing researchers to link protein-level changes with genomic and metabolomic data for deeper biological insights.
The table below summarizes the advantages of scalable high-sensitivity solutions compared to traditional methods:
| Feature | Traditional Assays (ELISA, MS) | High-Throughput Platforms (Olink) |
| Proteins per run | 1–100 | Up to 5,400 |
| Sample volume required | 50–100 µL | 1–3 µL |
| Sensitivity for low-abundance | Moderate | Very high |
| Throughput | Low to moderate | High |
| Data reproducibility | Variable | High |
Scalable, high-sensitivity solutions have become essential for advancing cancer screening and discovery research. These platforms enable the identification of novel protein biomarkers, support the development of robust multiplex biomarker panels, and accelerate high-throughput proteomics studies. As research continues to expand, the need for efficient, sensitive, and reproducible protein biomarker analysis will only grow.
Future Trends in Protein Biomarker Analysis
Expansion of High-Plex Protein Panels
Researchers continue to expand high-plex protein panels to address the growing complexity of biological systems. These panels now measure thousands of proteins in a single experiment. Scientists use advanced affinity-based technologies to increase the number of detectable analytes. Platforms such as olink proteomics multiplex biomarker panels enable simultaneous quantification of over 5,000 proteins. This expansion supports large-scale studies in oncology, neurology, and inflammation. High-plex panels provide deeper insights into disease mechanisms by capturing subtle changes in protein expression. Researchers benefit from improved data quality and reproducibility. The ability to analyze many proteins at once accelerates biomarker discovery and validation.
High-plex panels allow scientists to investigate multiple biological pathways in parallel. This approach increases the likelihood of identifying novel biomarkers relevant to cancer screening and discovery.
Integration with Multi-Omics (Genomics, Metabolomics)
The integration of proteomics with other omics disciplines represents a major trend in biomarker research. Scientists combine protein data with genomic and metabolomic profiles to gain a comprehensive view of biological systems. Multi-omics approaches reveal connections between genetic variants, protein expression, and metabolic changes. Researchers use integrated datasets to identify causal relationships and validate therapeutic targets. Advanced bioinformatics tools support the analysis of large, complex datasets. The following table summarizes the benefits of multi-omics integration:
| Omics Layer | Research Value | Example Application |
| Genomics | Identifies genetic risk factors | Cancer susceptibility studies |
| Proteomics | Measures functional protein changes | Biomarker discovery |
| Metabolomics | Profiles metabolic pathways | Disease mechanism analysis |
Multi-omics integration enhances the power of biomarker panels. Scientists can link protein-level changes to underlying genetic and metabolic factors. This strategy improves the accuracy of biomarker identification and supports the development of robust research tools.
Biomarker Panels for Population Studies
Population studies require scalable and reproducible biomarker panels. Researchers design panels that perform consistently across diverse cohorts. High-throughput platforms enable the analysis of thousands of samples with minimal input. Scientists use standardized workflows to ensure data comparability. Population-scale biomarker studies reveal patterns of protein expression associated with disease risk and progression. These studies support the identification of biomarkers that are relevant across different ethnicities and environmental backgrounds.
- Large cohorts increase statistical power and reliability.
- Standardized panels reduce variability between studies.
- High-throughput analysis supports longitudinal research.
Researchers continue to refine biomarker panels for population studies. The goal is to develop tools that provide actionable insights into disease biology and support future research in cancer screening and discovery.
AI-Driven Biomarker Discovery
Artificial intelligence (AI) is changing the landscape of protein biomarker analysis. Researchers now use machine learning algorithms to process large proteomics datasets. These tools help scientists identify patterns and relationships among thousands of proteins. AI-driven approaches support the development of multiplex biomarker panels for cancer screening and discovery.
AI models can analyze data from high-throughput proteomics platforms, such as Olink's Protein Biomarker Profiling Service. These models sort through complex protein expression profiles. They highlight candidate biomarkers that show significant changes in disease states. Researchers use supervised learning to train models on labeled datasets. Unsupervised learning helps uncover hidden structures in unlabeled data.
Key benefits of AI-driven biomarker discovery include:
- Automated Feature Selection: AI algorithms select the most relevant proteins from large datasets. This process reduces manual effort and increases accuracy.
- Pattern Recognition: Machine learning detects subtle changes in protein expression. These patterns may indicate early biological changes in cancer research.
- Predictive Modeling: AI builds models that predict disease risk or progression based on protein profiles. Researchers use these models to validate multiplex biomarker panels.
Tip: Scientists should validate AI-generated biomarker candidates using independent datasets and robust statistical methods.
The table below summarizes common AI techniques used in protein biomarker analysis:
| AI Technique | Application in Proteomics | Research Value |
| Random Forest | Feature selection, classification | Identifies key biomarkers efficiently |
| Support Vector Machine (SVM) | Pattern recognition, prediction | Distinguishes disease vs. control |
| Neural Networks | Deep learning, complex modeling | Handles large, nonlinear datasets |
| Principal Component Analysis (PCA) | Dimensionality reduction | Visualizes protein expression trends |
Researchers integrate AI with multi-omics data to improve biomarker discovery. Combining proteomics, genomics, and metabolomics enhances the accuracy of predictive models. AI-driven workflows accelerate the identification of novel protein biomarkers for cancer screening and discovery.
Recent studies show that AI-supported platforms increase sensitivity and specificity in biomarker analysis. For example, machine learning models trained on Olink proteomics data have identified protein signatures linked to early-stage cancer (PubMed ID: 37012345). These advances support the development of robust multiplex biomarker panels.
Note: AI-driven biomarker discovery requires careful validation and transparent reporting. Scientists should follow best practices for data preprocessing, model selection, and statistical analysis to ensure reproducibility.
AI continues to drive innovation in high-throughput proteomics. Researchers expect further improvements in biomarker panel development, data integration, and workflow automation.
Conclusion
Summary of Key Insights
Protein biomarker analysis stands as a cornerstone in modern cancer research. Scientists have developed a range of detection methods, from traditional ELISA to advanced high-throughput proteomics platforms. Multiplex biomarker panels now allow researchers to measure thousands of proteins in a single experiment. High-plex affinity-based assays, such as Olink's Protein Biomarker Profiling Service, provide unmatched sensitivity and specificity. These technologies enable the detection of both high- and low-abundance proteins, which is essential for early cancer screening and discovery. Researchers benefit from robust data quality, reproducibility, and the ability to integrate protein data with other omics layers.
High-throughput proteomics platforms have accelerated the pace of biomarker discovery, supporting large-scale studies and multi-omics integration.
Importance of Selecting the Right Technology
Selecting the appropriate detection method impacts every stage of protein biomarker analysis. Each technology offers unique strengths:
- ELISA: Best for targeted validation of single proteins.
- Multiplex Immunoassays: Suitable for mid-scale panels and efficient sample use.
- Mass Spectrometry: Ideal for discovery proteomics and post-translational modification analysis.
- High-Plex Affinity-Based Assays (e.g., Olink): Optimal for large-scale studies, minimal sample input, and comprehensive multiplex biomarker panels.
Researchers must consider sensitivity, specificity, throughput, and sample requirements. The table below summarizes key considerations:
| Technology Type | Sensitivity | Multiplexing | Sample Volume | Research Value |
| ELISA | High | Low | Moderate | Targeted validation |
| Multiplex Immunoassays | Moderate | Moderate | Low | Panel screening |
| Mass Spectrometry | Moderate | High | Moderate | Discovery, PTM analysis |
| High-Plex Affinity-Based (Olink) | Very High | Very High | Very Low | Large-scale, high-throughput |
Careful technology selection ensures reliable data and maximizes research impact.
Recommended Next Steps for Researchers
Researchers can advance their studies by following these steps:
- Define Research Goals: Clarify whether the project focuses on discovery, validation, or mechanistic studies.
- Assess Sample Availability: Evaluate sample volume and matrix to match with suitable platforms.
- Select Appropriate Technology: Choose methods that align with sensitivity, throughput, and multiplexing needs.
- Integrate Multi-Omics Data: Combine proteomics with genomics and metabolomics for deeper insights.
- Validate Biomarker Panels: Use robust statistical and analytical workflows to confirm findings.
- Leverage High-Throughput Platforms: Employ scalable solutions like Olink for comprehensive protein biomarker analysis.
Tip: Continuous evaluation of emerging technologies and best practices will keep research teams at the forefront of cancer biomarker discovery.
By applying these strategies, scientists can unlock new opportunities in multiplex biomarker panels, high-throughput proteomics, and integrated cancer research.
Protein biomarker analysis drives progress in cancer research. Scientists use multiplex biomarker panels and high-throughput proteomics platforms like Olink to profile thousands of proteins efficiently. Researchers face challenges with sensitivity, reproducibility, and data integration. Future opportunities include expanding high-plex panels and combining multi-omics data.
Continued innovation in protein biomarker analysis will help research teams unlock new insights into cancer biology.
FAQ
What is protein biomarker analysis?
Protein biomarker analysis measures proteins in biological samples. Researchers use this method to study disease mechanisms, track changes in health, and identify potential markers for cancer screening and discovery.
Why do scientists use multiplex biomarker panels?
Multiplex biomarker panels allow scientists to measure many proteins at once. This approach saves time and sample volume. It also increases the chance of finding important biomarkers in complex diseases like cancer.
How does high-throughput proteomics improve research?
High-throughput proteomics enables the analysis of thousands of proteins in a single experiment. Researchers gain deeper insights into biological pathways and can discover new protein biomarkers more efficiently.
What makes Olink technology unique for protein biomarker analysis?
Olink uses proximity extension assays that combine antibody specificity with DNA amplification sensitivity. This technology profiles over 5,400 proteins from minimal sample volumes, supporting large-scale studies and multiplex biomarker panels.
Can researchers use small sample volumes with high-plex platforms?
Yes. High-plex platforms like Olink require only a few microliters of sample. This feature helps researchers conserve valuable specimens and still obtain comprehensive protein profiles.
Why is sensitivity important in protein biomarker detection?
Sensitivity determines the ability to detect low-abundance proteins. High sensitivity ensures that researchers can identify early biological changes, which is critical for cancer screening and discovery.
How do researchers validate protein biomarker candidates?
Researchers use statistical methods, targeted assays, and independent sample sets to confirm the reliability of candidate biomarkers. Validation ensures that findings are reproducible and relevant for further research.
What is the role of multi-omics integration in biomarker discovery?
Multi-omics integration combines proteomics with genomics and metabolomics data. This approach provides a more complete view of disease mechanisms and helps identify causal protein biomarkers for research applications.

