Overview
Neurodegenerative and psychiatric disorders represent one of the greatest challenges in modern medicine, with neurological conditions now surpassing cardiovascular diseases as the leading cause of global disease burden. Innovative proteomic technologies are revolutionizing our understanding of these conditions and creating new possibilities for diagnosis and treatment. The Olink Neurology panel, based on Proximity Extension Assay (PEA) technology, has emerged as a particularly powerful tool for neuroscientists and translational researchers. This article explores the diverse applications of this cutting-edge platform in neurological research, with special emphasis on Alzheimer's disease, Parkinson's disease, and psychiatric disorders, while examining the protein signaling pathways revealed by this technology.
Olink PEA Technology: A Key to Unlocking Proteomics in Neuroscience Research
1. Innovative Technology and Exceptional Sensitivity: The Olink platform employs an innovative method called PEA, which integrates antibody-based immunoassays with PCR amplification. This technology achieves exceptional sensitivity, capable of detecting proteins in the fg/mL range, which is crucial for identifying low-abundance proteins that are often key biomarkers in neurological diseases.
2. Minute Sample Volume Requirement: A key advantage of PEA technology is its ability to perform simultaneous quantification of multiple proteins from very small sample volumes, requiring as little as 1μl. This effectively addresses a major limitation of traditional proteomic methods, especially when working with precious samples like cerebrospinal fluid (CSF) in neurological research.
3. Specialized Neurology Panels for Targeted Research: Olink offers strategically designed, neurology-specific panels (such as the Target 96 Neurology and Neuro Exploratory panels). These panels target proteins relevant to neurological processes and disorders, including established markers for conditions like Alzheimer's and Parkinson's disease, as well as exploratory proteins involved in broader biological processes such as neurodevelopment, axonal guidance, synaptic function, and immune regulation. This design allows researchers to investigate specific hypotheses while remaining open to new discoveries.
4. Bridging Peripheral and Central Nervous System Compartments: A significant advantage of this platform for neurological disorder research is its capability to correlate protein levels between peripheral fluids (like plasma) and the central nervous system (using CSF). This suggests that blood-based biomarkers could provide insights into brain pathophysiology, which is particularly valuable for longitudinal studies and clinical trials where repeated CSF collection is impractical or unethical.
5. Technical Robustness and Reproducibility for Large-Scale Studies: The platform demonstrates strong technical robustness, generating consistent and reproducible results across different sample types, including plasma, serum, and CSF. This consistency is essential for the reliability of large-scale, multicenter studies, which are common in neurodegenerative disease research.
Unraveling Alzheimer's Disease: Biomarker Discovery and Pathway Analysis
Alzheimer's disease (AD) research has been transformed by proteomic profiling approaches, with Olink technology playing a pivotal role in identifying novel biomarkers and signaling pathways. The panel's comprehensive coverage includes proteins directly relevant to AD pathogenesis, such as those involved in amyloid-beta metabolism, tau pathology, neuroinflammation, and synaptic dysfunction.
Early Protein Signatures
A recent study published in Nature Neuroscience utilized Olink proteomics technology to analyze cerebrospinal fluid samples from the BioFINDER-2 cohort. This research identified 127 differentially abundant proteins associated with the progression of Alzheimer's disease pathology, revealing distinct signatures such as SMOC1 upregulation in early Aβ plaque stages and proteins like MAPT and NRGN linked to tau tangle accumulation. The study demonstrated that Olink technology uncovers staged proteomic alterations in AD, offering biomarkers for early intervention and insights into disease-specific pathways.
 Figure 1: Differential protein abundance in CSF between AT comparisons. (Pichet Binette, A. et al., 2024)
Figure 1: Differential protein abundance in CSF between AT comparisons. (Pichet Binette, A. et al., 2024)
Protein Pathways and Mechanisms
A large-scale proteomic study published in Molecular Psychiatry investigated the relationship between inflammatory dietary patterns and plasma proteins to identify molecular mediators of future cognitive impairment. Analyzing data from 1,528 cognitively unimpaired women in the Women's Health Initiative Memory Study (WHIMS), researchers used the Energy-Adjusted Dietary Inflammatory Index (EDII) to quantify dietary inflammation and Olink proteomics to measure 151 inflammatory/immune proteins. Over a 14-year follow-up, they identified seven plasma proteins—CXCL10, CCL3, HGF, OPG, CDCP1, NFATC3, and ITGA11—that were not only associated with pro-inflammatory diets but also predicted incident cognitive impairment, with external validation in independent cohorts (ESTHER and ARIC).
This study underscores how pro-inflammatory diets modulate specific plasma proteins to influence cognitive health, offering novel biomarkers for early detection and potential targets for dietary interventions.
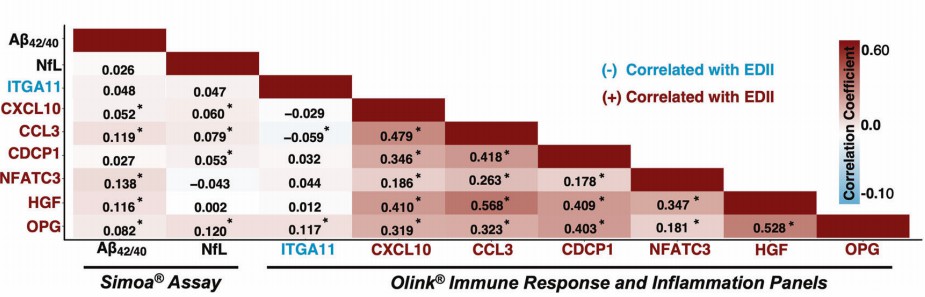 Figure 2: Heatmap showing correlations and corresponding coefficients between candidate proteins and plasma biomarkers of brain amyloid-β (Aβ42/40) and neurodegeneration (NfL). (Duggan, M.R. et al., 2023)
Figure 2: Heatmap showing correlations and corresponding coefficients between candidate proteins and plasma biomarkers of brain amyloid-β (Aβ42/40) and neurodegeneration (NfL). (Duggan, M.R. et al., 2023)
Decoding Parkinson's Disease: Molecular Subtypes and Therapeutic Targets
Parkinson's disease (PD) research has similarly benefited from the application of Olink proteomic panels, with recent studies revealing intriguing insights into disease heterogeneity and progression. The multiplexing capability of the Olink platform is especially valuable in PD, where the clinical presentation is highly variable and the underlying pathophysiology extends beyond the classical dopamine system.
Genetic Risk and Protein Expression
A large-scale proteomic study published in Nature Medicine leveraged plasma samples from the Global Neurodegeneration Proteomics Consortium (GNPC) to delineate shared and disease-specific molecular pathways across Alzheimer's disease (AD), Parkinson's disease (PD), and frontotemporal dementia (FTD).
This study provides a comprehensive plasma proteomic atlas of neurodegeneration, revealing conserved and distinct pathways that enhance understanding of disease mechanisms. The findings support the utility of plasma-based biomarkers for precision diagnostics and identify novel targets for therapy.
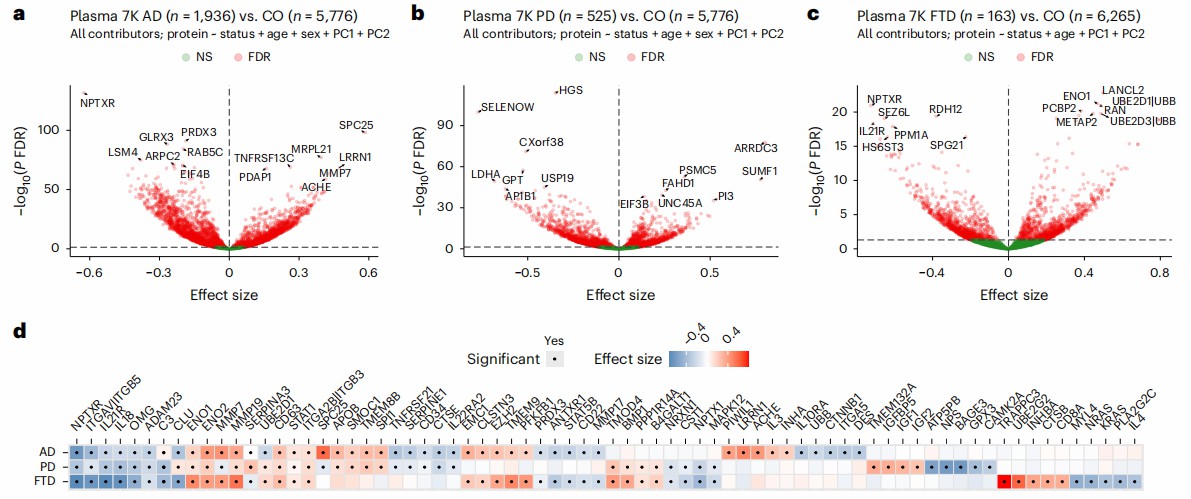 Figure 3: Plasma proteomic alterations in AD, PD and FTD compared to controls. (Ali, M. et al., 2025)
Figure 3: Plasma proteomic alterations in AD, PD and FTD compared to controls. (Ali, M. et al., 2025)
Protein Dynamics and Disease Progression
The recent introduction of Olink's Target 48 neurodegenerative panel represents a strategic response to the need for focused assays in Parkinson's research. This panel includes 41 proteins specifically associated with neurodegenerative diseases, with optimization for plasma detection.
The integration of proteomic data with other molecular data types provides a more comprehensive view of Parkinson's pathogenesis. A multi-omics approach combining proteomic, genomic, transcriptomic, and metabolomic data can elucidate the complex interplay between different pathological processes in Parkinson's, including neuroinflammation, mitochondrial dysfunction, and protein aggregation.
Psychiatric Disorders: Bridging the Gap Between Immunology and Neuroscience
Neuro-Immune Axis in Schizophrenia and Depression
Olink profiling has uncovered significant inflammatory protein dysregulation in psychiatric disorders. For instance, an analysis of neuron-derived exosomes in HIV-associated neurocognitive impairment revealed sex-specific patterns: men showed increases in proteins like carboxypeptidase M, while women had decreases in proteins such as cathepsin S, echoing sex differences observed in Alzheimer's disease. (Sun, B. et al., 2019)
These findings support the emerging hypothesis that immune system dysfunction plays a causal role in certain psychiatric conditions, potentially opening new therapeutic avenues targeting specific inflammatory pathways. The identification of proteins with transdiagnostic relevance could lead to therapeutic strategies targeting shared pathways, potentially offering benefits across multiple conditions.
Protein Biomarkers as Objective Measures
A study employs the Olink Proseek Multiplex Platform to conduct large-scale proteomic profiling of plasma samples from World Trade Center responders, identifying protein biomarkers associated with post-traumatic stress disorder (PTSD), mild cognitive impairment (MCI), and their comorbidity. The research revealed 50 differentially expressed proteins, with multiprotein composite scores achieving high diagnostic accuracy (AUC up to 0.84), highlighting the potential of Olink-based proteomics to provide objective measures for classifying and monitoring psychiatric and cognitive conditions.
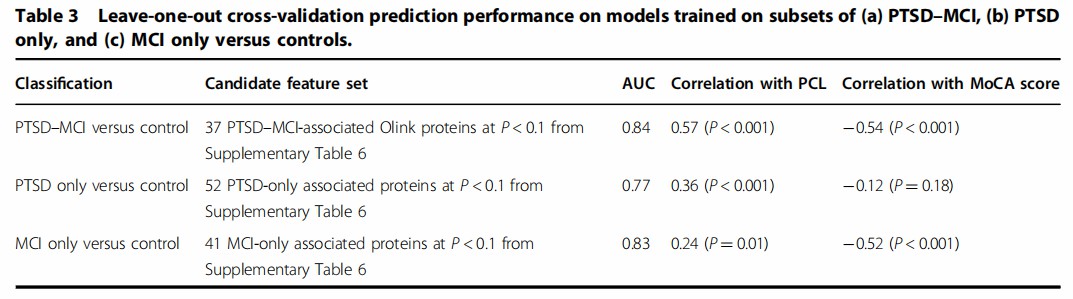 Figure 4: Predictive performance of multiprotein composite scores derived from Olink Proteomic data in differentiating psychiatric and cognitive conditions. (Kuan, PF. et al., 2020)
Figure 4: Predictive performance of multiprotein composite scores derived from Olink Proteomic data in differentiating psychiatric and cognitive conditions. (Kuan, PF. et al., 2020)
Advancing Drug Development
The integration of Olink proteomic data with genomic information through Mendelian randomization approaches provides a powerful framework for target validation. This methodology uses genetic variants as instrumental variables to assess causal relationships between protein levels and disease risk. This approach increases confidence in potential targets by reducing confounding and reverse causation, major limitations of observational studies.
For example:
- A study used large-scale CSF protein analysis to identify 31 protein groups linked to frontotemporal dementia (FTLD). Key findings include increased activity in RNA splicing pathways (especially in genetic carriers) and decreased synaptic function, both correlating with disease severity. These results were confirmed in independent cohorts, highlighting proteins like NPTX2 as promising biomarkers. (Saloner, R. et al., 2025)
- Another study introduces a minimally invasive diagnostic method for glioma by analyzing plasma extracellular vesicles (plEVs) with high-sensitivity immunoprofiling. Using techniques like size exclusion chromatography for isolation and Olink's proximity extension assay for protein detection, the research identifies syndecan-1 (SDC1) as a reliable biomarker. (Indira Chandran, V. et al., 2019)
Conclusion and Future Directions
- Key Advantages of Olink Neurology Panel
High sensitivity, high-plex capability, and compatibility with low-volume samples (e.g., CSF) enable robust proteomic profiling in neurological and psychiatric research. - Emerging Trends in Olink Applications
(1) Expansion of protein panels (covering thousands of proteins) allows hypothesis-free discovery and deeper proteome coverage.
(2) Integration with multi-omics (genomics, transcriptomics, metabolomics) reveals shared/disease-specific pathways. - Future Directions
(1) Proteomic signatures may lead to point-of-care assays for early diagnosis and monitoring.
(2) Combining proteomic data with digital biomarkers improves disease subtyping and prognosis prediction.
(3) Early detection of neurological conditions years before clinical onset opens avenues for preventive interventions.
Olink technology advances understanding of neurological/psychiatric disorders, with growing potential for precise diagnostics and therapeutics through scalable proteomic and multi-omics integration.
FAQs
1. What are the key advantages of using Olink compared to traditional immunoassays?
Key advantages include:
- Sensitivity: Detects proteins at sub-pg/mL concentrations.
- Multiplexing: Simultaneously analyzes 92 biomarkers in a single run.
- Sample Efficiency: Requires only 1–10 μL of biofluids (CSF, plasma).
- Standardization: Provides normalized protein expression (NPX) values for cross-study comparisons.
2. Can Olink data be combined with genomics or transcriptomics?
Yes. Common integration strategies include:
- Mendelian Randomization: Linking protein QTLs to disease risk.
- Pathway Enrichment: Correlating protein modules with transcriptomic networks.
- Machine Learning: Building multi-omic diagnostic classifiers.
3. What are the current limitations of Olink technology?
Limitations include:
- Fixed panel design (limited customizability vs. untargeted mass spectrometry)
- Lower protein coverage than discovery proteomics
- Requiring validation via orthogonal methods (e.g., ELISA)
4. What sample size is recommended for cohort studies?
For cohort studies utilizing the Olink platform, sample size recommendations vary based on the primary research objective: for initial biomarker discovery, a pilot study typically requires 50–100 samples per group, while validation studies demand larger cohorts of at least 200 per group to ensure statistical robustness; for more complex aims such as clinical subtyping or machine learning-based classification, a total sample size of 500 or more is generally recommended to account for multivariate analysis and model stability.
Related Reading
For deeper insights, we recommend exploring the following related guides:
Olink Proteomics in Clinical Research: A Game Changer — An overview of how Olink's proteomics platform is revolutionizing clinical research by enhancing diagnostic accuracy and accelerating personalized medicine through high-throughput protein biomarker validation.
Olink Proteomics in Alzheimer's Disease Research — An overview of how Olink's proteomics platform is advancing Alzheimer's disease research by identifying disease-specific proteins, revealing pathological mechanisms, and enabling precision medicine through large-scale CSF and plasma proteomics.
References
- Pichet Binette, A., Gaiteri, C., Wennström, M. et al. Proteomic changes in Alzheimer's disease associated with progressive Aβ plaque and tau tangle pathologies. Nat Neurosci 27, 1880–1891 (2024).
- Duggan, M.R., Butler, L., Peng, Z. et al. Plasma proteins related to inflammatory diet predict future cognitive impairment. Mol Psychiatry 28, 1599–1609 (2023).
- Ali, M., Erabadda, B., Chen, Y. et al. Shared and disease-specific pathways in frontotemporal dementia and Alzheimer's and Parkinson's diseases. Nat Med 31, 2567–2577 (2025).
- Olink® Target 48 Neurodegeneration
- Sun, B., Fernandes, N., Pulliam, L. et al. Profile of neuronal exosomes in HIV cognitive impairment exposes sex differences. AIDS 33, 1683-1692 (2019).
- Kuan, PF., Clouston, S., Yang, X. et al. Molecular linkage between post-traumatic stress disorder and cognitive impairment: a targeted proteomics study of World Trade Center responders. Transl Psychiatry 10, 269 (2020).
- Saloner, R., Staffaroni, A.M., Dammer, E.B. et al. Large-scale network analysis of the cerebrospinal fluid proteome identifies molecular signatures of frontotemporal lobar degeneration. Nat Aging 5, 1143–1158 (2025).
- Indira Chandran, V., Welinder, C. et al. Ultrasensitive Immunoprofiling of Plasma Extracellular Vesicles Identifies Syndecan-1 as a Potential Tool for Minimally Invasive Diagnosis of Glioma. Clin. Cancer Res. 25, 3115-3127 (2019).



