Multi-omics datasets can look complete yet still fail to guide translational decisions. Teams often discover compelling RNA signatures, only to watch candidates stall when protein evidence, timing, or cohort variability tells a different story. This guide is designed to help PIs and systems biology teams make auditable, decision-ready choices by integrating Olink proteomics with genomics and transcriptomics—especially in clinical cohort protein profiling.
If you're unfamiliar with PEA technology or NPX data, start with the Olink workflow overview: Olink Proteomics Assay Services Explained: From PEA Technology to Research-Ready Data.
Key takeaways
- Protein-level evidence should anchor decisions where pathway activity, druggability, and immune state matter; RNA is discovery-friendly but rarely sufficient for validation on its own.
- In clinical cohort protein profiling, plan synchronous sampling or tightly staged designs to minimize timing-driven discordance between RNA and protein.
- Define a decision hierarchy up front: specify when protein leads, when RNA supports mechanism, and what orthogonal checks are required before advancing.
- Design for harmonization: include bridge samples, document QC thresholds, and apply robust batch correction (bridging → empirical Bayes or mixed models).
- Use immune and inflammatory pathways as an integration stress test; if signals cohere under immune dynamics, your pipeline is likely robust.
The Role of Protein-Level Evidence in Clinical Cohort Biomarker Decisions
Transcriptomics excels at discovery—broad coverage, sensitivity to transcriptional changes, and rich pathway annotations. But validation requires confidence that molecular signals translate into functional differences. Protein abundance sits closer to pathway activity, receptor–ligand interactions, and circulating immune responses. That proximity makes proteomics the practical anchor for translational decisions.
- Pathway activity and druggability: Proteins are effectors and drug targets. A transcript increase without a corresponding protein change may indicate post-transcriptional control or short-lived RNA without functional impact.
- Immune state and systemic response: Many cytokines and chemokines are secreted and dynamic. Circulating protein profiles capture the organism-level reaction more directly than tissue RNA alone.
- Clinical cohort protein profiling benefits: When cohorts are multi-center or longitudinal, protein-level readouts provide decision-grade anchors to interpret genetic and RNA signals, helping triage which candidates merit validation.
Authoritative analyses consistently show modest mRNA–protein concordance with frequent discordance driven by regulation and stability. For example, cross-tissue studies report only limited alignment between layers and highlight tissue-specific disagreement, underscoring why protein evidence must be weighed heavily in translational choices; see tissue-wide concordance assessments in peer-reviewed cross-layer studies (2019–2023) and mechanistic explorations of discordance in Nucleic Acids Research (2023).
Where Genomics, Transcriptomics, and Proteomics Agree — and Where They Don't
Agreement and discordance between layers are both informative. The key is to translate patterns into decision rules.
Concordant signals: when RNA and protein move together
Concordance (e.g., increased RNA and increased protein) strengthens confidence that a pathway is engaged and that a candidate biomarker may be actionable. In validation:
- Use concordant signals to justify advancing to orthogonal assays (e.g., ELISA or targeted MS) and clinical stratification tests.
- Confirm that concordance persists across centers and timepoints; require consistency under batch correction and <LOD handling.
- Map concordant proteins to target accessibility and pharmacodynamics; if the protein is druggable and concordant with RNA, validation odds improve.
Discordant signals: when RNA and protein tell different stories
Discordance is common and often mechanistic rather than an error:
- Post-transcriptional regulation and translation control (RBPs, miRNAs) can mute protein changes despite RNA shifts; see systematic analyses summarized in Journal of Cell Biology (2023).
- Secreted proteins and circulating markers may spike rapidly and decay, while tissue RNA reflects production capacity rather than immediate systemic levels.
- Immune proteins are transient and context-dependent, frequently diverging from RNA due to rapid signaling cascades.
Decision rules when discordant:
- If the clinical endpoint depends on circulating activity (e.g., cytokine-mediated inflammation), prioritize protein evidence for decisions and use RNA for mechanism mapping.
- If discordance points to timing mismatches, adjust sampling windows or add longitudinal points before advancing.
- Require orthogonal validation when layers diverge; do not advance candidates on RNA alone without protein support.
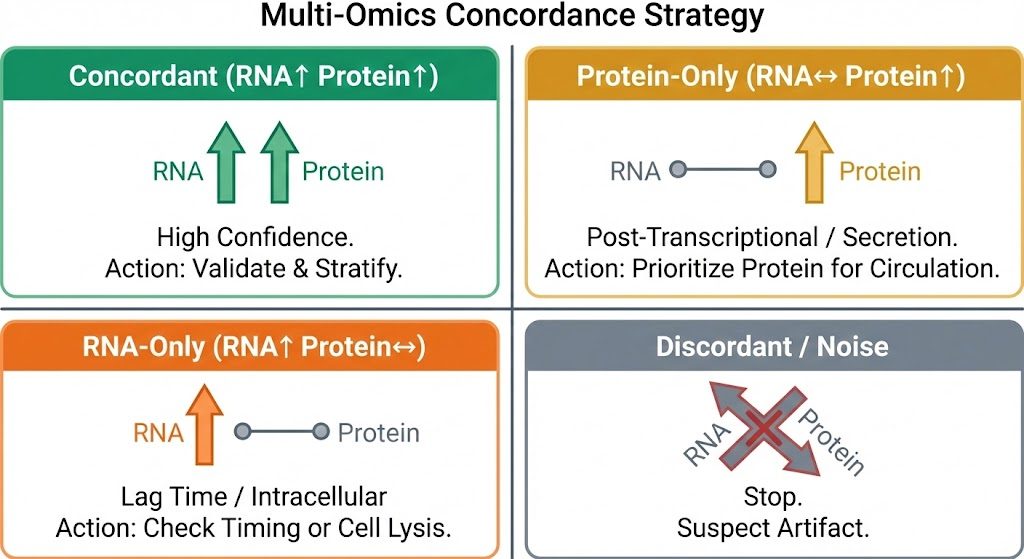 Decision framework for interpreting RNA–protein concordance.
Decision framework for interpreting RNA–protein concordance.
For deeper context on discordance causes and implications, see integrative analyses in Nucleic Acids Research (2023) and cohort-level summaries in open-access cross-tissue datasets (2023).
Practical Framework: Integrating Olink Proteomics into Multi-Omics Study Design
When to introduce proteomics in a multi-omics pipeline
- Synchronous sampling: Collect genomics/transcriptomics/proteomics at the same visit for robust cross-layer correlation and causal modeling. This reduces timing-driven discordance, particularly for immune signals.
- Staged introduction: Start with RNA/WGS discovery, then add targeted proteomics for triage and validation. Use this when budgets are tight, but mitigate risk with carefully matched sampling windows.
- Protein-anchored design: In plasma/serum cohorts, anchor decisions on protein-level effects and use RNA to explain mechanism and cell-of-origin.
Cohort considerations for integrated analysis
- Sample size and power: Base calculations on expected NPX effect sizes and variance; include site as a random effect and simulate minimum detectable fold changes under batch structure.
- Timepoints and longitudinal design: Align windows to biology (e.g., 1–2 weeks for cytokine shifts). Standardize collection time, minimize freeze–thaw cycles, and capture concomitant medications.
- Clinical phenotypes and stratification: Predefine strata (e.g., responders vs non-responders) and incorporate covariates to avoid confounding. Register decision thresholds to maintain auditability.
For a deeper dive into cohort and QC planning in large studies, see Designing a Large-Scale Proteomics Study with Olink: Cohorts, Power, and QC Strategy.
Creative Proteomics provides Olink proteomics assay services and multi-omics integration support. Mentioned here for context only; no promotional claims.
Immune and Inflammatory Pathways as a Multi-Omics Stress Test
Immune pathways are where multi-omics most frequently "argues," making them ideal for stress-testing integration quality.
- Rapid dynamics: Cytokines and chemokines change quickly in circulation; protein measurements capture systemic activity that tissue RNA may miss.
- Ligand–receptor networks: Integration with single-cell transcriptomics helps map circulating proteins to producing or responding cell types.
- Olink strengths: High analytical specificity of PEA enhances confidence when integrating secreted immune markers with RNA data; see analytical notes summarized by Illumina in the Olink proteomics technical overview (2024).
If you focus on immune-related translational research, read Olink Immune Profiling in Translational Research: From Discovery to Validation.
Common Integration Pitfalls — and How to Avoid Them
- Treating multi-omics as additive data rather than a decision tool: Predefine your hierarchy (protein leads for circulating and immune signals; RNA supports mechanism) and specify required orthogonal checks.
- Letting RNA dominate and relegating protein to post hoc validation: Triaging candidates on protein effect sizes after QC avoids late-stage failure.
- Ignoring batch, site, and time effects: Include bridge samples across plates/sites; apply bridging normalization; if needed, use empirical Bayes (ComBat) or linear mixed models to correct batch effects.
- Mishandling <LOD values: Document your approach (exclude, impute, or censored models) and perform sensitivity analyses.
- Underpowered cohorts: Simulate power for NPX changes with realistic variance; design longitudinal sampling aligned to biology to boost signal.
Recommended methods and resources:
- Bridging normalization best practices are introduced in the OlinkAnalyze bridging vignette and expanded in Olink's data normalization white paper.
- When bridging is limited, empirical Bayes via ComBat (sva) and linear mixed models for complex designs are reliable alternatives.
- Large-cohort precedent: UK Biobank integrated Olink proteomics across thousands of samples with subset normalization and extensive bridging; see the UKB Olink 1536 analysis report.
From Integrated Data to Actionable Biomarker Decisions
Advance, hold, or stop decisions should be explicit and reproducible.
Advance when:
- Protein effects are robust after QC/bridging and consistent across centers and timepoints.
- Signals align with pathway mechanisms (e.g., ligand–receptor logic) and, ideally, with genetic anchors (cis/trans pQTL or colocalization with GWAS).
- Orthogonal assays confirm magnitude/direction; early stratification improves model fit.
Hold when:
- RNA effects lack protein support or appear timing-dependent.
- Batch correction materially changes effect estimates; <LOD handling dominates conclusions.
- Evidence conflicts across matrices (e.g., plasma vs CSF) without a mechanistic explanation.
Stop when:
- Protein signals fail replication under harmonized analysis.
- Discordance suggests non-functional transcriptional noise or technical artifact.
- Antibody cross-reactivity is suspected and not resolved by orthogonal checks.
Creative Proteomics offers Olink-based multi‑omics integration services. For teams seeking operational support—panel selection, bridging‑sample design, NPX/QC reporting, and integrated analysis workflows—see Creative Proteomics' multi‑omics integration page for an example engagement model: Creative Proteomics multi‑omics integration.
FAQ: Multi-Omics Integration with Olink Proteomics
When should proteomics be added to a genomics or transcriptomics study?
Introduce proteomics at discovery for broad profiling or after RNA discovery for targeted triage. In clinical cohorts, a protein-anchored design often improves translational relevance by capturing pathway activity and circulating immune signals.
How should discordant RNA and protein signals be interpreted?
Treat discordance as a mechanistic clue: consider post-transcriptional control, secretion, and timing. Prioritize protein for functional decisions, and use RNA to map mechanism and cell-of-origin. Validate with orthogonal assays before advancing.
Is Olink proteomics suitable for large clinical cohorts?
Yes. Olink's Explore platforms support high throughput and minimal sample volumes, and large cohorts (e.g., UK Biobank) have demonstrated robust normalization and integration across thousands of samples.
Can immune-related proteins improve multi-omics biomarker validation?
Often. Secreted cytokines and chemokines reflect systemic activity and therapy response. Pairing Olink proteomics with single-cell RNA can map signals to producing cell types, strengthening validation.
What are common reasons multi-omics biomarkers fail in validation?
Unaddressed batch/site/time effects, timing mismatches between RNA and protein sampling, mishandled <LOD values, and using multi-omics as additive data rather than a decision framework.
Conclusion: Making Multi-Omics Work for Translational Decisions
The value of multi-omics is not "more data"; it is fewer wrong decisions. Protein-level evidence—especially in clinical cohort protein profiling—translates molecular signals into clinical relevance. By planning sampling windows, defining decision hierarchies, designing for harmonization, and stress-testing integration in immune pathways, you can reduce validation failure and advance only those biomarkers with decision-grade support.
FAQ: Multi-Omics Integration with Olink Proteomics
Q: When should proteomics be added to a genomics or transcriptomics study?
A: Add proteomics either at discovery (broad profiling) or after RNA discovery for targeted triage; in clinical cohorts where translational decisions depend on circulating biology, a protein‑anchored design improves decision relevance. For large-cohort operational details, see the guide on designing large-scale Olink studies (cohorts, power, QC).
Q: How should discordant RNA and protein signals be interpreted?
A: Treat discordance as a mechanistic clue: evaluate post‑transcriptional regulation, secretion/clearance, and sampling timing; prioritize protein evidence for circulating or immune-dependent endpoints and require orthogonal validation (targeted MS or ELISA) before advancing.
Q: How should bridging samples be designed for cross‑center Olink studies?
A: Use a panel of biological bridging samples (10–30 per batch depending on throughput) that span the NPX dynamic range and represent cohort diversity; randomize their placement across plates, include pooled duplicates, and predefine acceptance criteria. Verify adjustment effectiveness with PCA/variance decomposition and replicate concordance before accepting corrected NPX (see the OlinkAnalyze bridging vignette for implementation details).
Q: What is a practical rule for handling values below LOD in NPX data?
A: Pre-specify a handling strategy: exclude assays with excessive <LOD (for example >50%), apply censored regression or substitution (LOD/√2) for sparse <LOD, and run sensitivity analyses to confirm conclusions are robust to the choice.
Q: When should teams use empirical Bayes (ComBat) versus linear mixed models for batch correction?
A: Use ComBat when batches are reasonably balanced and biological covariates can be modeled in the design matrix; prefer linear mixed models when designs are unbalanced, have nested random effects (site/plate), or when preserving sample‑level contrasts is critical—validate results by comparing biological effect sizes after correction (see ComBat documentation for technical details).
Q: Can genetic evidence (pQTL) help resolve RNA–protein discordance?
A: Yes. Cis/trans pQTLs that colocalize with trait GWAS or other genetic anchors strengthen confidence in a protein signal even when RNA is discordant; use genetic colocalization as a complementary mechanistic anchor, not as a sole determinant.




