Proteomics is evolving fast. The rise of Olink proteomics technology, especially its Olink Cytokine panels and high-plex Olink panels, is helping to bridge historic gaps in sensitivity, throughput, and clarity. As researchers push toward single-cell resolution, spatial mapping, integrated omics, and AI-driven insights, we must ask: where does Olink stand now, and how can it be leveraged in future applications?
Emerging Frontiers in Proteomics Research
The proteomics field in 2025 is being reshaped by multiple converging trends. Here are the key ones:
Population-scale proteomics: Large cohorts are becoming central. For example, the UK Biobank Pharma Proteomics Project (UKB-PPP) is using Olink's Explore HT platform to target over 5,400 proteins across 600,000 samples.
Multi-ancestry, multi-omics integration: Proteogenomics, combining protein expression data with genetic, transcriptomic, phenotypic data, is revealing deeper insights into genetic control of protein levels, disease risk, and biomarker discovery.
Single-cell and spatial proteomics: Technologies that allow protein quantification at the level of individual cells or within spatially preserved tissue sections are maturing. Challenges (sample prep, sensitivity, data analysis) remain serious but progress is rapid.
AI, machine learning, and computational advances: With datasets now reaching tens of thousands of samples and thousands of proteins (e.g. UKB-PPP's ~3,000 proteins in ~54,000 samples in pilot) researchers are developing new statistical models, ML pipelines, and federated learning frameworks to mine the data
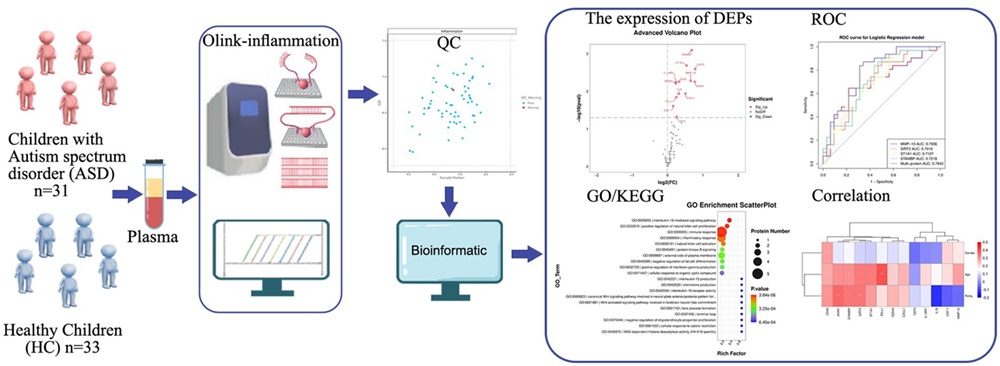 Fig 1. Workflow for plasma ASD vs healthy control comparison using the Olink Inflammation panel: QC → DEP detection → GO/KEGG enrichment.
Fig 1. Workflow for plasma ASD vs healthy control comparison using the Olink Inflammation panel: QC → DEP detection → GO/KEGG enrichment.
Olink's Unique Strengths in This Landscape
Given those emerging frontiers, Olink offers certain advantages that make it particularly well suited—and some areas to watch.
Core Advantages
High-Throughput with Defined Panels: Olink's Explore HT can measure thousands of predefined proteins in plasma or serum with good reproducibility. For example, the UKB-PPP pilot used Olink Explore 3072 to profile ~3,000 proteins in >54,000 participants.
Sensitive Detection of Low-Abundance Proteins: Many cytokines, growth factors, signaling molecules are present at very low concentrations. Olink's technology, especially the Olink Cytokine panels, allows detection of many of these with low sample volume.
Standardisation, Reproducibility, Multiplex Compatibility: Because the panels are well-defined, with consistent assay designs, batch correction tools, and robust QC pipelines (e.g. limit of detection, NPX data handling in UKB-PPP) researchers can trust cross-sample, cross-site comparisons.
Where Olink Excels vs Others
vs mass-spectrometry (MS): Olink is faster to run for large sample numbers, uses less sample prep for many fluid samples, and yields a cleaner output when you're looking for predefined proteins. MS has advantages in discovery of novel proteins, post-translational modifications, and in tissue sections.
vs other affinity‐based platforms (e.g. aptamer methods): Olink has robust antibody pairs and good specificity, and increasingly broad panels (especially with the newer HT Explore panels covering thousands of proteins). For example, comparative studies show that Olink has a high proportion of assays with cis-protein quantitative trait loci (pQTLs) supporting assay performance. In one UK Biobank / SomaScan comparison, 72% of Olink assays had supporting evidence vs ~43% for SomaScan for a shared set of assays.
Integration with Other Omics & AI-Driven Insights
To unlock the full potential of proteomics, integration with other omics and computational methods is essential.
Proteogenomics: The UKB-PPP work demonstrates this. With ~3,000 plasma proteins from over 54,000 participants, thousands of novel genotype-protein associations (pQTLs) were detected. Over 14,000 pQTLs identified; ~67% had cis associations, strengthening confidence in measurement reliability.
Machine Learning and Predictive Models: Researchers are using the large Olink datasets to build phenome-wide and disease risk models. Larger sample sizes help reduce overfitting and allow for more robust feature selection. Bootstrap methods, cross-validation across ancestries, and federated learning are becoming more common.
Handling Data Complexity: With many missing values (below limits of detection), batch effects, and heterogeneity in sample types, advanced normalization, imputation and QC pipelines matter. Olink datasets (e.g. UKB-PPP) provide resources like NPX values, batch IDs, LODs to help researchers apply correct filters.
This leads to better biological insights: pathway analyses, co-expression networks, causal inference (e.g. Mendelian randomization) become feasible at scale.
Spatial, Single-Cell, and In Vivo Proteomics — Can Olink Adapt?
As the field moves toward single-cell resolution and spatial context, the question is whether Olink's formats and panels can stretch to those demands.
Single-Cell Proteomics: Current single-cell techniques (mostly MS-based) are pushing towards detection of hundreds to thousands of proteins per cell (SCoPE2, etc.). Olink panels are not currently designed for single-cell resolution. The requirements (very low sample volume, high sensitivity) pose technical challenges.
Spatial Proteomics / In Situ Assays: Mapping protein expression within tissue architecture is increasingly important (e.g. tumor microenvironment, organ structure). Mass spectrometry imaging, multiplexed immunofluorescence / immunohistochemistry, and new spatial transcriptomics are active areas. Olink, being designed for fluid or homogenised samples, may need adaptation (new sample prep, miniaturisation, possibly microfluidics or spatial barcoding) if it seeks to address spatial proteomics.
In Vivo Sensing / Longitudinal Sampling: Longitudinal plasma sampling with Olink panels is already happening in cohort studies. But in vivo sensing (e.g. implants, microdialysis, sampling interstitial fluid) would demand robustness of the assay in new matrices, as well as low sample volumes and stable reagents.
In short, Olink is strong in bulk fluid proteomics, population scale, and pre-defined panels; to compete or integrate with spatial and single-cell frontiers, further innovation is needed.
Key Challenges & Opportunities for Olink in Future Proteomics
As with any technology, Olink has advantages — but also constraints. Recognising both helps researchers use it well, and helps developers anticipate directions.
Challenges
Plex Depth & Novel Targets: Even with Explore HT targeting thousands of proteins, the panels are limited to predefined sets. Discovering previously unmeasured proteins or rare isoforms still often falls to MS-based discovery.
Sample Diversity: Most data are from serum/plasma. Sample types such as tissue biopsies, exosomes, interstitial fluid, or FFPE specimens pose additional challenges (matrix effects, antigen retrieval, signal loss).
Dynamic Range and Sensitivity: Detecting ultra-low abundance proteins remains hard, especially in complex mixtures. Multiplexing sometimes comes with trade-offs in sensitivity.
Cost, Turn-Around Time, and Scalability: As sample size scales (e.g. 50,000+), costs of reagents, consumables, logistic QC etc. become nontrivial. Also, lab throughput, data storage, computational requirements all scale up.
Standardisation and Protocol Harmonisation: Cross-platform comparisons (Olink vs SomaScan vs MS) show differences in which proteins are reliably measured, limit of detection, and signal linearity. These differences must be well understood to avoid misinterpretation. The UKB comparison highlighted that while number of pQTLs was similar, the proportion of assays with supporting cis associations was much higher for Olink.
Opportunities
Large Cohort & Longitudinal Studies: The UKB-PPP and similar efforts show the power of massive datasets. Academic institutions can plan studies with Olink panels from the start, ensuring sample collection, processing, and QC align.
Hybrid Omics Packages: Researchers combining proteomics, transcriptomics, metabolomics, epigenomics will find value in integrating Olink data. Linking with panels like How to Integrate Olink Proteomics with Other Omics Technologies helps. (Internal link to Article No. 14)
Cytokine / Immune Profiling in High Plex: Given increasing interest in immunology, inflammation, immune signatures, Olink Cytokine panels remain strong assets. Combining with spatial or cellular resolution may be possible.
Cloud / AI-First Analytical Tools: Providing more accessible pipelines, dashboards, annotation tools (e.g. phenotype-to-proteome mapping, biomarker candidate ranking) will help non-computational labs.
Enhanced Panels & Reagents: Developing adaptions for new sample matrices, improving sensitivity, expanding dynamic range, and possibly moving toward newer modalities like in situ or barcoded PEA reagents.
Where Olink Fits in Oncology, Biomarker Discovery, Inflammation & Immune Research
To ground this in concrete applications, here are areas where Olink's strengths map particularly well.
Oncology & Biomarker Discovery: Large-scale plasma proteomics can lead to biomarkers for cancer risk, progression, and treatment response. UKB-PPP data already supports many such studies. (Olink Proteomics in Oncology and Biomarker Discovery)
Immune System / Inflammation Research: Olink Cytokine panels are particularly powerful in profiling immune signaling, inflammation pathways, cytokine storms, immune regulation. As the need grows for multiparameter immune signatures, combining Olink cytokine data with single-cell or spatial immune mapping could yield new insights.
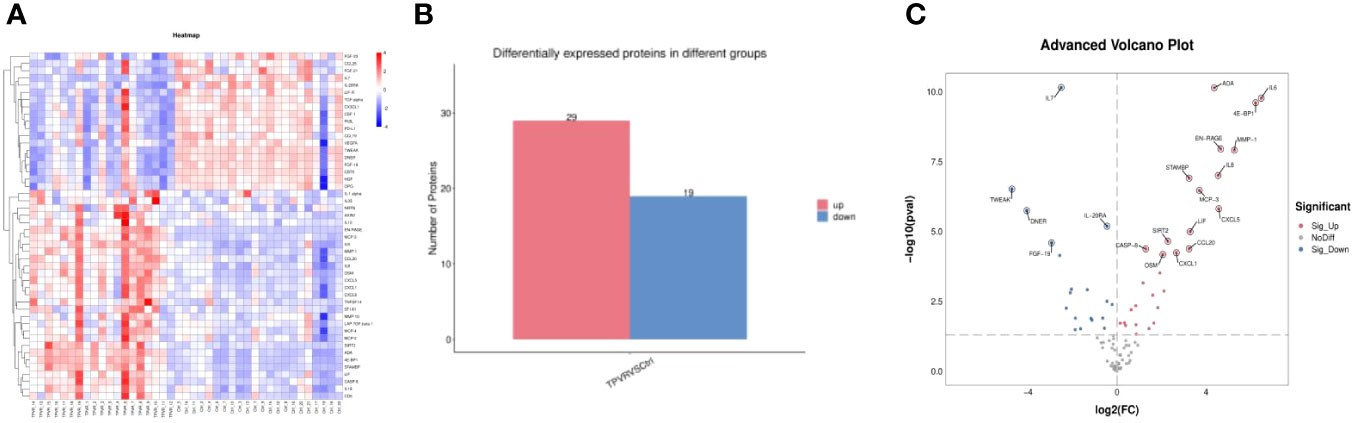 Fig 2. Heatmap and volcano plot showing inflammation-related proteins: up- and down-regulated markers via Olink inflammation panel.
Fig 2. Heatmap and volcano plot showing inflammation-related proteins: up- and down-regulated markers via Olink inflammation panel.
Longitudinal and Population Studies: Tracking proteomic changes over time in large cohorts (ageing, lifestyle, exposures) provides evidence for causal inference and prediction. UKB work using Olink Explore panels is already delivering in this realm.
Rare Variant / Genetic Association Studies: Because Olink panels show good pQTL support (especially cis associations), they are useful in genetic epidemiology where linking genotype to protein expression strengthens downstream functional inferences.
Strategies for Researchers — How to Leverage Olink Going Forward
If your lab or institution plans to integrate Olink into cutting-edge proteomics workflows, here are practical strategies.
Design Studies with Scale and QC in Mind
- Define sample types early (plasma vs serum, tissue, etc.), plan for batch effects, and collect metadata (e.g. sample handling, storage).
- Pilot small subset: test detection limits, verify antibodies / panels.
Choose the Right Panel(s)
- Use Olink Cytokine or inflammation panels for immune signaling and inflammation questions.
- For discovery or broad coverage, select Olink Explore HT panels.
- Consider cost vs coverage trade-offs.
Combine with Other Omics (Multi-Modal Studies)
- Integrate proteomics with transcriptomics and genomics for proteogenomics insights. (How to Integrate Olink Proteomics with Other Omics Technologies)
- Use ML/AI pipelines to integrate large proteomic datasets with phenotype, genotype, and, where possible, imaging or spatial data.
Data Handling, Normalisation, and Interpretation
- Filter for proteins exceeding limit of detection; correct for batch/plate effects using metadata. UKB datasets provide LODs, plate IDs.
- Use imputation or appropriate statistical approaches for missing data.
- Validate key findings using orthogonal methods (ELISA, MS) when feasible.
Invest in Future-Ready Infrastructure
- Sample storage, lab automation, computational pipelines, cloud resources.
- Collaborations with core facilities or CROs if your in-house throughput is limited.
Conclusion
Olink proteomics technology is already deeply embedded in the future of proteomics. With its strengths in measuring thousands of proteins in large cohorts, detecting low-abundance cytokines, and offering reproducible multiplex panels, it serves well for many non-clinical research applications. Yet, to fully align with emerging needs — single-cell resolution, spatial context, novel targets — further innovation and thoughtful study design will matter.
If you're considering incorporating Olink Cytokine panels or broader Olink panels into your next project:
Start by exploring Introduction to Olink Proteomics: What You Need to Know o ensure foundational alignment.
We invite you to contact us for custom study planning using Olink panels. Whether you need help selecting panels, designing QC pipelines, or integrating proteomics into a broader omics framework, our team of experts is ready to support your research journey.
References
- Sun et al., 2023. "New biological truths discovered in the world's largest proteogenomic study."
- "High-throughput proteomics: a methodological mini-review," Olink, (2022)
- Rushdy Ahmad & Bogdan Budnik, "A review of the current state of the art of single-cell proteomics and future perspective," Anal. & Bioanal. Chem., 2023
- Grimur Hjorleifsson Eldjarn et al., 2023. Large-scale plasma proteomics comparisons through genetics and disease associationsBao et al., "Olink proteomics profiling platform reveals non-invasive inflammatory related protein biomarkers in autism spectrum disorder", Frontiers in Molecular Neuroscience, 2023; DOI: 10.3389/fnmol.2023.1185021
- Guo H, Wang T, Yu J, Shi Z, Liang M, Chen S, He T, Yan H. Vitreous Olink proteomics reveals inflammatory biomarkers for diagnosis and prognosis of traumatic proliferative vitreoretinopathy. Front Immunol. 2024 Feb 22;15:1355314. doi: 10.3389/fimmu.2024.1355314. PMID: 38455059; PMCID: PMC10917961.








