Introduction
In the world of proteomics, researchers are constantly searching for technologies that offer high sensitivity, accuracy, and scalability. With the growing demand for reliable and reproducible results, traditional proteomics methods can sometimes fall short, particularly when it comes to analyzing large datasets or identifying low-abundance proteins. The need for precision, especially in complex biological samples, is now more important than ever.
This is where Olink Proteomics steps in, offering a revolutionary solution that transforms the landscape of proteomics research. Olink's cutting-edge platform allows for accurate, multiplex protein analysis with unmatched sensitivity, providing researchers with valuable insights that drive scientific discovery.
What if you could measure up to 92 proteins in a single sample—simultaneously, with high accuracy and minimal sample volume? And what if you could do this with a technology that scales easily from small-scale studies to large cohort analyses, all while reducing time and cost?
This is not just a possibility—it's a reality with Olink Proteomics.
If you're curious about how to choose the best Olink panel for your study, see our article "How to Choose the Right Olink Panel for Your Research.
What Is Olink Proteomics?
Olink Proteomics is a groundbreaking technology designed to meet the growing demands of proteomics research. By leveraging its unique Proximity Extension Assay (PEA) technology, Olink offers highly sensitive and precise measurements of up to 92 proteins in a single run, all from a minimal sample volume. This allows researchers to gain deeper insights into protein function and interactions, significantly enhancing their ability to conduct high-quality studies.
The main advantage of Olink proteomics is its ability to provide highly reproducible results with minimal variation between samples, even in complex biological matrices like blood or plasma. Traditional methods, such as Western blotting or mass spectrometry, often struggle with reproducibility and require large amounts of sample material, which can be a limitation in studies where sample availability is scarce.
Olink's multiplexing capability sets it apart from traditional proteomics methods. Researchers can analyze a broad range of proteins simultaneously, which not only saves valuable time but also reduces costs. The ability to process large numbers of samples efficiently allows for high-throughput analysis, making Olink ideal for large cohort studies and clinical research applications.
In addition, Olink's platform is scalable and flexible, catering to various research needs from small-scale exploratory studies to large epidemiological investigations. Whether you are studying biomarkers for disease, immune responses, or cellular pathways, Olink provides a versatile solution that can meet the diverse needs of proteomics research.
- If you need a quick comparison of 96-plex versus 48-plex formats, please see "Olink 96 vs 48-Plex Panels: Key Differences".
The Science Behind Olink
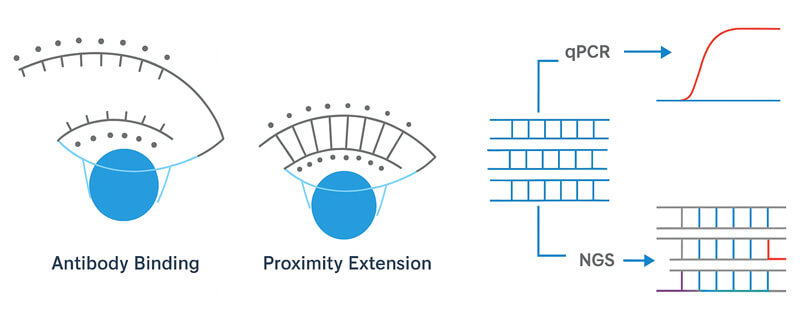
Olink's Proximity Extension Assay (PEA) technology is a groundbreaking advancement in proteomics, offering unparalleled specificity and scalability for high-throughput protein analysis. At its core, PEA employs a dual-antibody system, where each antibody is conjugated with a unique DNA oligonucleotide. When these antibodies bind to their respective epitopes on the target protein, the proximity of the DNA tags facilitates hybridization, forming a double-stranded DNA molecule. This DNA product is then amplified using quantitative PCR (qPCR) or next-generation sequencing (NGS), providing a robust and sensitive readout of protein abundance.
One of the standout features of PEA is its exceptional specificity. The requirement for two antibodies to bind simultaneously to the same protein significantly reduces the likelihood of cross-reactivity, a common challenge in multiplexed assays. Studies have demonstrated that Olink's PEA technology achieves an analytical specificity of 99.7%, ensuring that the detected signals accurately correspond to the target proteins.
This high specificity is complemented by the platform's scalability. Olink's panels can measure up to 92 proteins in a single sample, with the Olink Explore platform capable of profiling over 5,400 proteins across thousands of samples simultaneously. Such scalability is invaluable for large-scale studies, enabling comprehensive proteomic profiling without compromising data quality.
Moreover, the integration of PEA with advanced data analysis tools enhances the reliability and interpretability of the results. Olink's proprietary NPX Manager software offers rigorous quality control measures and statistical analyses, ensuring that the data generated are both accurate and reproducible.
In summary, Olink's PEA technology represents a significant leap forward in proteomics, combining high specificity, scalability, and advanced data analysis to provide researchers with a powerful tool for protein biomarker discovery and validation.
- To understand throughput options and when to consider Explore/HT runs, please see "Olink's High-Throughput Proteomics: What It Means for Your Research".
Comparing Olink to Alternative Methods (MS, ELISA, Aptamer Platforms)
Why this section matters. Researchers often weigh Olink's Proximity Extension Assay (PEA) against mass spectrometry (MS), classical ELISA, and aptamer-based assays (e.g., SomaScan). The studies below show these methods are complementary rather than interchangeable, and they differ in coverage, specificity, dynamic range, sample volume, and genetics-anchored validity.
Olink (PEA) vs Mass Spectrometry (LC-MS/MS)
Design & readout. Olink is an affinity-based, targeted assay using dual antibodies with DNA tags (PEA) read by qPCR/NGS, enabling high-sensitivity multiplexing from minimal volume. MS is untargeted/system-wide, excels at proteoform/PTM discovery, but typically needs more input and instrument time. In side-by-side work, teams highlight their complementarity—PEA for sensitive, efficient quantification across many samples; MS for broad discovery and proteoform detail.
When to prefer each. Choose Olink for low-abundance biomarkers across large cohorts or longitudinal studies; use MS to characterize isoforms/PTMs or discover novel proteins before moving to targeted validation.
Olink (PEA) vs Single-plex/Low-plex Immunoassays (ELISA)
Multiplexing & volume. ELISA remains robust for single-analyte quantification but scales poorly to tens–hundreds of proteins. Olink routinely profiles dozens to thousands with stringent internal controls, preserving sample for additional work. Comparative overviews describe ELISA as confirmatory and Olink as an efficient discovery/validation bridge in research-use-only workflows. Specificity. PEA requires dual-recognition events (two antibodies), reducing off-target signal—helpful in complex matrices where cross-reactivity can confound single-plex assays.
Olink (PEA) vs Aptamer Platforms (e.g., SomaScan)
Correlation & genetic anchors. Large head-to-head studies show only modest correlation between matched protein measurements across Olink and SomaScan, reflecting platform-specific binding/detection biology. Notably, Olink proteins are more likely to have cis-pQTLs (genetic variants near the encoding gene that affect protein levels), supporting target specificity; where both platforms have cis-pQTLs, many colocalize, underscoring complementarity rather than contradiction. Complementary disease insights. Integrative analyses using both platforms have revealed non-overlapping signals that, when combined, strengthen biological interpretation and disease mapping—an argument for multi-platform strategies in population-scale studies.
Practical selection guide (research-use-only)
High-throughput cohort profiling (minimal sample, many proteins): favor Olink PEA; consider orthogonal MS or aptamer follow-up when you need broader coverage or to reconcile assay-specific effects.
Discovery of novel isoforms/PTMs: start with MS, then port key candidates to Olink (if available) or other targeted assays for scalable validation.
Cross-platform robustness: where feasible, triangulate with genetics (pQTLs) and replicate hits across Olink and aptamer platforms to increase confidence in causal signals.
For more details on how Olink's cytokine panel performs against MSD and Luminex platforms in throughput, sensitivity, and sample volume, see the article "Olink Cytokine Panel vs Other Cytokine Detection Methods"
How Olink's Cytokine Panel Accelerates Inflammation Research
Inflammation is a fundamental biological response to infection, injury, or harmful stimuli, playing a crucial role in various physiological and pathological processes. Understanding the intricate network of cytokines—small signaling proteins that mediate and regulate immunity and inflammation—is essential for advancing research in immunology, oncology, and autoimmune disorders.
Olink's Cytokine Panel offers a comprehensive solution for profiling multiple cytokines simultaneously with high sensitivity and specificity. Utilizing the Proximity Extension Assay (PEA) technology, the panel enables the measurement of up to 92 cytokines from a single sample, providing a broad overview of the inflammatory landscape.
One of the key advantages of Olink's Cytokine Panel is its exceptional specificity. The dual-antibody recognition system inherent in PEA technology ensures that only true binding events contribute to the signal, minimizing cross-reactivity and background noise. This high specificity is crucial when analyzing complex biological samples where low-abundance cytokines may be present.
Moreover, the scalability of Olink's platform allows researchers to expand their studies beyond cytokines to include other protein biomarkers, facilitating a more holistic understanding of inflammatory pathways. Whether investigating the role of cytokines in disease progression, treatment response, or biomarker discovery, Olink's Cytokine Panel provides a robust and reliable tool for advancing inflammation research.
- For a plain-English overview of assay advantages and where cytokine panels fit, please see "Olink's Cytokine Assays: Key Benefits and Applications in Research".
- For immune-system use cases and experimental patterns, please review "Using Olink's Cytokine Panels for Immune System Research".
Case Study: Using Olink Proteomics to Investigate Cancer Biomarkers
Olink Proteomics has been instrumental in advancing cancer biomarker research by enabling the simultaneous analysis of multiple proteins with high sensitivity and specificity. This capability is particularly valuable in oncology, where understanding the complex interplay of proteins can lead to the identification of novel biomarkers for early detection, prognosis, and therapeutic targets.
For instance, a study published in Frontiers in Immunology utilized the Olink Inflammation Panel to analyze the inflammatory protein profiles in patients with traumatic proliferative vitreoretinopathy (TPVR). The researchers identified several differentially expressed proteins that could serve as potential biomarkers for TPVR progression, demonstrating the utility of Olink's technology in identifying disease-specific biomarkers.
Similarly, another study employed the Olink Target 96 Inflammation Panel to profile serum proteins in patients with Kawasaki Disease (KD). The analysis revealed elevated levels of IL-17A, CCL23, and other cytokines, which were associated with the disease's inflammatory response. These findings underscore the potential of Olink Proteomics in uncovering inflammatory pathways relevant to cancer and other diseases.
Furthermore, Olink's technology has been applied in neuroinflammation research, where it was used to profile cerebrospinal fluid samples from patients with Guillain-Barré Syndrome (GBS). The study identified specific inflammatory mediators that could aid in the diagnosis and understanding of GBS pathogenesis, highlighting the versatility of Olink Proteomics across different biological contexts.
These case studies illustrate the broad applicability of Olink Proteomics in cancer biomarker discovery and beyond. By providing high-throughput, multiplexed protein analysis, Olink enables researchers to gain deeper insights into the molecular underpinnings of diseases, facilitating the identification of novel biomarkers and therapeutic targets.
- For a broader strategy on discovery, verification, and translation in oncology, please see "Olink Proteomics in Oncology and Biomarker Discovery".
- If your datasets include serum or plasma and you need help with result reading, please review "How to Interpret Results from Olink's Serum Proteomics Panels".
Olink's Plasma Proteomics: Unlocking Insights for Disease Research
Plasma proteomics offers a powerful approach to understanding complex diseases by analyzing the protein content in blood plasma. Olink's Proximity Extension Assay (PEA) technology enables the measurement of thousands of proteins simultaneously with high sensitivity and specificity, making it a valuable tool in disease research.
Case Study: Alzheimer's Disease
A comprehensive study utilizing Olink's Target 96 panels profiled nearly 1,200 plasma proteins in individuals with Alzheimer's disease (AD) and healthy controls. The research identified a 19-protein signature that accurately distinguished clinical AD, with an area under the curve (AUC) ranging from 0.969 to 0.9816. These findings not only provide a foundation for developing a high-performance, blood-based test for AD screening and monitoring but also offer abundant protein targets for future therapeutic development.
Case Study: Type 2 Diabetes and Prediabetes
In a study conducted with the Białystok PLUS Cohort, plasma samples from individuals with newly diagnosed type 2 diabetes and prediabetes were analyzed using Olink Reveal. The analysis revealed proteins significantly associated with glucose tolerance, HbA1c levels, and glycemia during oral glucose tolerance tests. Notably, BAIAP2 was identified as a protein consistently associated with hyperglycemia across all time points, suggesting its potential as a marker for early-stage diabetes.
Case Study: Parkinson's Disease
A large-scale, multi-tissue proteomics study aimed at identifying biomarkers for early diagnosis and disease monitoring in Parkinson's disease (PD) utilized Olink's Target 96 panels. The study identified several proteins, including DOPA decarboxylase (DDC), consistently upregulated in the cerebrospinal fluid and urine of treatment-naïve PD patients. These findings suggest that DDC could serve as a promising diagnostic and prognostic marker for PD.
Case Study: Sepsis
In critically ill sepsis patients, targeted plasma proteomics using Olink's Target 96 panels identified 25 proteins on ICU Day-1 and 26 proteins on ICU Day-3 that distinguished sepsis patients from healthy controls with ≥90% classification accuracy. These proteins were correlated with various clinical and laboratory variables, providing pathophysiological insights into sepsis and potential biomarkers for patient stratification.
- If you need guidance on interpreting multi-analyte serum outputs, please read "How to Interpret Results from Olink's Serum Proteomics Panels"
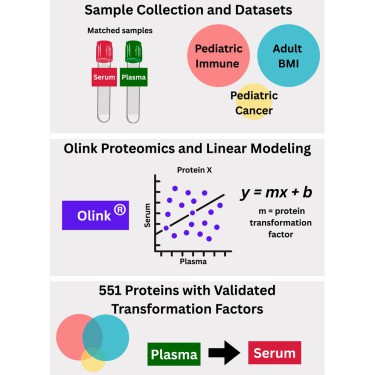 Proximity extension assay (PEA), have enabled high-throughput multiplex protein analyses from small sample volumes in either serum or plasma.
Proximity extension assay (PEA), have enabled high-throughput multiplex protein analyses from small sample volumes in either serum or plasma.
How to Use Olink's Data for Translational Research: Key Insights
Olink proteomics bridges basic discovery and translational research by offering deep proteomic profiling that informs downstream validation and target development. Its unique combination of high specificity and scalability makes it ideal for uncovering biological mechanisms and informing experimental design.
A key example is the integration of Olink proteomic data with transcriptomic profiling to dissect cellular responses. In a recent study, researchers applied Olink-targeted panels alongside transcriptomics to examine how TNF-α impacts mesenchymal stem cells (MSCs). They found that TNF-α induces cellular aging and programmed cell death while impairing stem cell migration and differentiation—insights that help clarify mechanisms underpinning tissue regeneration (Zhou et al., 2025).
Olink proteomics also supports large-scale biomarker discovery and target validation. For instance, the UK Biobank Pharma Proteomics Project uses Olink Explore to characterize over 5,400 proteins across hundreds of thousands of samples—data from which have already enabled early detection of cancer risk years before diagnosis.
Additionally, combining proteomic data with genetic information through Mendelian randomization studies helps identify causal disease markers. One such proteogenomic study used Olink PEA data to map circulating protein variations to genetic loci, offering causal insights that can inform drug target discovery.
These examples illustrate how Olink's rich, high-throughput datasets often drive translational pipelines—from mechanistic research in cell models to population-scale biomarker discovery and integrative multi-omics analysis.
- For oncology-specific pipelines and target qualification, please read "Olink Proteomics in Oncology and Biomarker Discovery".
Understanding Olink's Data Analysis Process: From Raw Data to Insights
Olink proteomics generates data as Normalized Protein eXpression (NPX) values, a relative quantification unit on a log₂ scale. NPX balances sensitivity and consistency, enabling comparisons of the same protein across samples within the same project.
- Quality Control (QC) is embedded in the workflow using a layered system of built-in internal and external controls:
- Internal Controls—such as Incubation, Extension, and Detection Controls—monitor each stage of the Proximity Extension Assay (PEA): binding, extension, and detection.
- External Controls—Negative Controls, Plate or Sample Controls, and Calibrators—are included per plate to assess assay performance, set limits of detection (LOD), and enable normalization across plates.
QC thresholds include:
- Run-level checks: Standard deviation of control NPX values should remain within 0.2 (samples) to 0.5 (external controls); exceeding this flags a QC issue.
- Sample-level checks: Control NPX values deviating more than 0.3 from the plate median trigger a sample QC warning.
After QC, data undergo normalization:
- Intensity Normalization is applied across fully randomized, multi-plate projects. It aligns medians of assays across plates to an overall median, eliminating systematic bias.
- In less randomized studies, Reference Sample Bridging is used. Shared samples across plates serve as anchors to adjust for batch effects.
- For large-scale and multi-project analyses, bridging becomes essential. Shared samples allow datasets to be aligned using batch adjustment in tools like the OlinkAnalyze R package (e.g., olink_normalization_n).
- Olink's software ecosystem, including NPX Map, NPX Manager, Analyze, and Insight, supports streamlined QC procedures, normalization, and visualization workflows—ensuring raw data becomes actionable insight.
- If you are formalizing SOPs for sample QC, plate controls, and bridging, please see "How to Set Up Olink Proteomics for Your Lab: A Step-by-Step Guide".
- For a step-by-step view of how NPX data are processed and validated, please read "Understanding Olink's Data Analysis Process: From Raw Data to Insights"
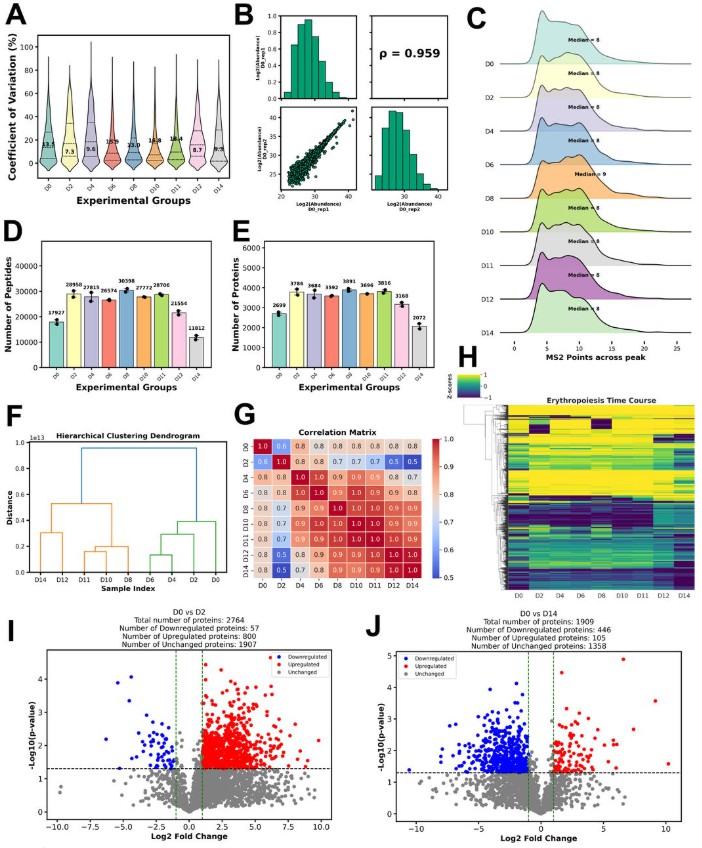
It illustrates concrete evidence of how proteomics data vary across conditions / time, giving readers a real use case of advanced data processing and visualization.
Population-Scale Proteogenomics: Olink at the Forefront of Large-Cohort Insights
The UK Biobank Pharma Proteomics Project (UKB-PPP) stands as the world's largest human proteomics study. Using the Olink Explore platform, researchers measured ~3,000 proteins across more than 54,000 UK Biobank samples. This remarkable dataset enabled researchers to map over 14,000 protein quantitative trait loci (pQTLs), 81% of which were new discoveries. These data are now serving as a powerful resource for causal inference in population-scale studies.
Crucially, these pQTLs support Mendelian randomization, linking protein levels to disease phenotypes and informing potential drug targets.
Building on these findings, the project is now scaling further. The UKB-PPP will employ Olink Explore to analyze 5,400+ proteins from up to 600,000 samples, making it the most comprehensive proteomics mapping initiative to date.
Studies have already demonstrated how proteogenomic integration—combining proteomics with whole-exome or genome sequencing—can advance drug discovery by uncovering novel biomarkers and accelerating translational research.
If you want to examine a concrete example of large-cohort execution, please read "Olink Proteomics for Large-Scale Population Studies: A Success Story"
Practical Insights: Olink in Inflammation Research
Olink proteomics excels at profiling inflammation-related proteins in diverse biological samples. For instance, a study utilizing the Olink Target 92 Inflammation Panel assessed serum from patients with compartment syndrome versus healthy controls. Significant elevations in IL-6, CSF-1, and HGF were found and confirmed by ELISA, showcasing Olink's reliability in detecting key inflammatory mediators (Fisher et al., 2022).
Another example leveraged the Olink Explore 1536 platform, enabling quantitative analysis of nearly 1,472 proteins—including those involved in neurology, oncology, metabolism, and inflammation—in the blood plasma of COVID-19 patients (unnamed study) . The data were processed and quality-controlled using Olink's NPX Manager software—a vital tool for ensuring data integrity.
Epidemiological research also benefits from Olink's technology. In an extensive study involving over 6,600 participants, researchers used the Olink Target 96 Inflammation Panel to profile inflammatory proteins and derive an "obesity-related inflammatory protein signature." This signature correlated strongly with mortality and cardiovascular risk—outperforming established genetic risk scores (Argentieri et al., 2024) .
- For a feature-level explanation of the inflammation panel used in many studies, please see "Exploring the Olink Inflammation Panel: Key Features & Applications".
- If you want to review end-to-end plasma workflows connected to inflammation profiling, please read "Olink's Plasma Proteomics: Unlocking Insights for Disease Research".
Take the Next Step with Olink Proteomics
Ready to elevate your proteomics research with precision, depth, and efficiency? Let's make it happen together.
Why choose us?
Accelerate your project with trusted Olink services, managed end-to-end from panel selection to advanced data analysis.
Access consultative support tailored to your needs, whether you're exploring discovery panels or embarking on translational studies.
Our collaborations have empowered researchers across institutions — offering scalable solutions that deliver high sensitivity from minimal sample volumes.
Take action now:
Contact our specialists to discuss your research goals and receive expert guidance.
Request a quote or schedule a discovery call — see how Olink aligns with your project timelines and budget.
Prefer to explore first? Download our panel overview guide to find fast answers.
We're here to support your discovery journey with leading Olink technology — from high-throughput profiling to precision analysis.
References
- Pietzner, M., Wheeler, E., Carrasco-Zanini, J. Synergistic insights into human health from aptamer- and antibody-based proteomic profiling. Nature Communications 12, 6822 (2021)
- Wang, B., Pozarickij, A., Mazidi, M. Comparative studies of 2168 plasma proteins measured by two affinity-based platforms in 4000 Chinese adults. Nature Communications 16, 1869 (2025)
- Eldjarn, G. H., Ferkingstad, E., Lund, S. H. Large-scale plasma proteomics comparisons through genetics and disease associations. Nature 622, 348–358 (2023)
- Shraim R, Diorio C, Canna SW, Macdonald-Dunlop E, Bassiri H, Martinez Z, Mälarstig A, Abbaspour A, Teachey DT, Lindell RB, Behrens EM. A Method for Comparing Proteins Measured in Serum and Plasma by Olink Proximity Extension Assay. Mol Cell Proteomics. 2025






