The Protein Quantification Challenge
The rapid advancement of genomic technologies has revolutionized our understanding of human biology and disease mechanisms. However, there remains a significant gap between genetic predisposition and phenotypic manifestation. This gap is largely filled by proteins as the primary functional executors of biological processes.
Despite their crucial role, comprehensive protein analysis has historically lagged behind genomic technologies due to formidable technical challenges. The immense dynamic range of protein concentrations in biological fluids (spanning over 10 orders of magnitude), the necessity for highly specific detection methods, and the complexity of protein structure and modifications have presented substantial obstacles to large-scale proteomic studies.
The Olink Explore 3072 platform represents a transformative approach to these challenges, leveraging innovative biochemistry and next-generation sequencing to achieve unprecedented specificity and multiplexing capability. This platform has positioned itself at the forefront of proteomic research by enabling simultaneous measurement of nearly 3,000 proteins from minimal sample volumes.
The PEA Technology Mechanism: Converting Protein Signals to DNA Barcodes
At the heart of the Olink Explore 3072 platform lies the Proximity Extension Assay (PEA) technology, an ingenious method that converts protein quantification into nucleic acid detection. This innovative approach combines the specificity of immunoassays with the sensitivity of PCR and scalability of next-generation sequencing.
The Stepwise PEA Process
The PEA technology operates through a precisely orchestrated sequence of molecular events:
- Dual Antibody Recognition: Each target protein is detected by a matched pair of antibodies, each conjugated with a unique DNA oligonucleotide tag. These antibodies are carefully selected or developed to bind to distinct epitopes on the target protein. When both antibodies simultaneously bind to their respective epitopes, their attached DNA oligonucleotides are brought into close proximity.
- Proximity-Driven DNA Hybridization: The spatial closeness of the two oligonucleotides enables them to hybridize, forming a double-stranded DNA template. This step is crucial for assay specificity—only when both antibodies correctly bind to their target protein do the DNA strands come sufficiently close to initiate hybridization. The requirement for dual recognition dramatically reduces non-specific signals that plague traditional immunoassays.
- Wide Dynamic Range: The platform achieves a quantitative range spanning over 10 orders of magnitude, capable of detecting both high-abundance proteins (e.g., albumin) and rare signaling molecules from the same sample.
- Reduced Batch Effects: The incorporation of internal controls and standardized normalization procedures (Normalized Protein eXpression, NPX) ensures data consistency across batches and studies, facilitating the combination of datasets from different sources and timepoints
 Figure 1: Modes of action of PEA probes for protein detection. (Dovgan, I. et al., 2019)
Figure 1: Modes of action of PEA probes for protein detection. (Dovgan, I. et al., 2019)
Table 1: Comparison of Proteomic Technologies
| Parameter | Traditional Immunoassays | Mass Spectrometry | Olink Explore 3072 |
| Multiplexing Capacity | Limited (typically<10-plex) | Moderate (hundreds) | High (3,000+ targets) |
| Sensitivity | Variable, moderate | Limited for low-abundance | fg/mL range |
| Sample Volume | Moderate to high | Moderate | Minimal (1-6μL) |
| Dynamic Range | 3-4 orders of magnitude | 4-5 orders | 10+ orders |
| Throughput | Low to moderate | Low | High (960 samples/run) |
Explore 3072 Platform Architecture: Modularity, Throughput, and Quality Control
The Olink Explore 3072 represents a sophisticated integration of biochemistry, automation, and data science specifically engineered for large-scale proteomic studies. Unlike conventional platforms that struggle to balance breadth and depth in protein measurement, this system employs a strategically partitioned approach to achieve comprehensive proteome coverage without compromising data quality.
Modular Panel Architecture
A distinctive architectural feature of the Explore 3072 platform is its organization into eight specialized 384-plex panels, each targeting proteins associated with specific biological domains:
- Cardiometabolic Panel: Focusing on markers relevant to metabolic syndrome, diabetes, and cardiovascular function
- Inflammation Panels: Comprehensive coverage of cytokines, chemokines, and inflammatory mediators
- Neurology Panels: Targeting proteins associated with neurodevelopment, neurodegeneration, and blood-brain barrier function
- Oncology Panels: Encompassing cancer-associated antigens, growth factors, and tumor microenvironment components
This modular configuration enables researchers to customize their analytical approach based on study objectives.
Automated Workflow and Quality Framework
The platform incorporates an automated workflow specifically designed for high-throughput operations, processing up to 960 samples in a single run. This scalability is essential for large biobank-scale studies such as the UK Biobank, where standardized processing and minimal technical variation are prerequisites for meaningful biological inference.
A cornerstone of the Olink platform is its rigorous quality control system, which operates at multiple levels:
- Sample-Specific Controls: Each reaction includes internal controls that monitor assay performance and correct for technical variability.
- Batch Normalization: Sophisticated normalization algorithms transform raw detection signals into Normalized Protein eXpression (NPX) values, enabling quantitative comparisons across samples and batches.
- Data Integration Protocols: Advanced bridging protocols facilitate the integration of new datasets with historical studies, extending the utility of existing proteomic resources.
This meticulous attention to quality control has established the platform as a benchmark for proteomic data reliability.
High-Throughput Data Capabilities: From Raw Signals to Biological Insights
The transition from raw protein measurements to biological understanding requires sophisticated data processing pipelines and analytical approaches. The Olink platform excels in this domain through its integrated bioinformatics ecosystem specifically designed for proteomic data.
NPX Normalization and Data Processing
The fundamental metric underlying Olink data analysis is the Normalized Protein eXpression (NPX) system, a standardized relative quantification unit that enables robust cross-sample comparisons. The NPX algorithm applies multiple correction factors to raw extension counts, accounting for technical variations while preserving biological signals. This normalized approach generates data that closely approximates log-normal distribution—a favorable characteristic for statistical modeling commonly employed in large-scale omics studies.
The Olink data processing pipeline encompasses several critical stages:
- Signal Normalization: Internal controls and sample-specific factors are used to calibrate protein measurements, reducing well-to-well and batch-to-batch variability.
- Quality Filtering: Samples and analytes failing quality thresholds are identified and excluded, with typical quality thresholds set to retain >85% of measured proteins in most sample types.
- Data Integration: Advanced algorithms facilitate the combination of datasets generated at different times or locations, enabling meta-analyses that amplify statistical power.
The platform's data output compatibility with standard bioinformatics tools allows researchers to apply a wide range of analytical approaches, from simple differential expression analysis to complex machine learning algorithms.
Exemplary Applications Across Biological Domains
The practical utility of the Olink Explore 3072 platform is exemplified through several landmark publications that have yielded novel biological insights:
- Lung Cancer Risk Prediction: Researchers identified protein signatures in blood samples taken up to 12 years before lung cancer diagnosis that predicted the disease with an AUC of 0.7. Different protein panels showed consistent predictive accuracy over time, indicating stable biological processes before clinical diagnosis. This long-term risk prediction could improve early detection for high-risk groups.
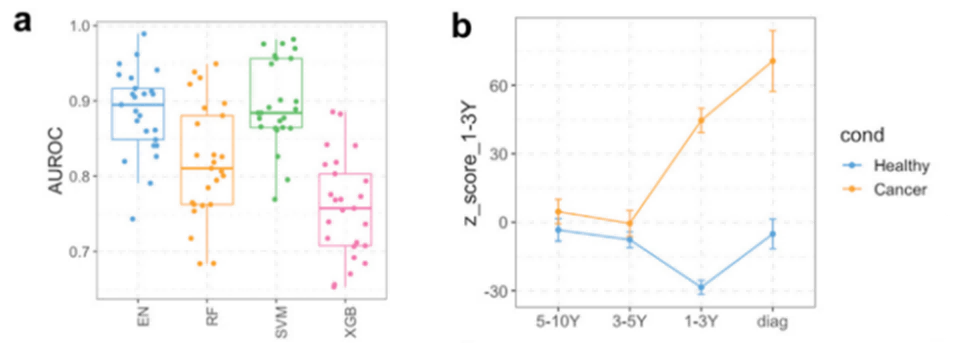 Figure 2: Olink Explore 3072 data shows machine learning models can accurately predict cancer risk as protein biomarker levels rise before diagnosis. (Michael, D. et al., 2023)
Figure 2: Olink Explore 3072 data shows machine learning models can accurately predict cancer risk as protein biomarker levels rise before diagnosis. (Michael, D. et al., 2023)
- Revolutionizing ALS Diagnosis: A study identified a 33-protein signature in blood that differentiates ALS patients from healthy individuals with 98.3% accuracy. The signature includes known markers such as neurofilament light chain (NFL) and novel proteins not previously linked to ALS. These protein changes were detectable years before clinical symptoms appeared, indicating potential for early disease detection.
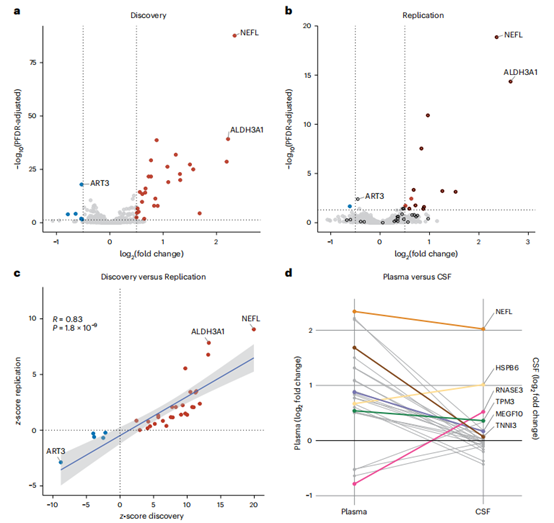 Figure 3: The Olink Explore 3072 analysis validates the consistency of key protein biomarkers across discovery, replication, and plasma versus CSF comparisons. (Chia, R. et al., 2025)
Figure 3: The Olink Explore 3072 analysis validates the consistency of key protein biomarkers across discovery, replication, and plasma versus CSF comparisons. (Chia, R. et al., 2025)
- Breast Cancer Causal Protein Discovery: researchers leveraged Olink's PEA Explore platform to perform high-throughput proteomic profiling of 2,929 unique proteins in plasma samples from 598 women in the KARMA cohort. The primary application was to identify circulating proteins with a potential causal role in breast cancer development using a Mendelian randomization (MR) framework.
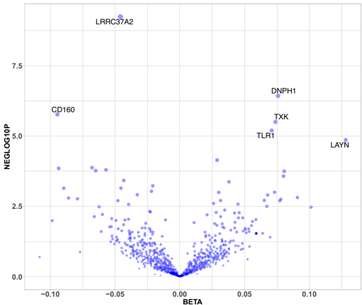 Figure 4: Volcano plot of effect sizes (X-axis) and −log10p (Y-axis) for the 730 proteins tested for breast cancer risk in the Mendelian randomisation analysis. (Mälarstig, A. et al., 2023)
Figure 4: Volcano plot of effect sizes (X-axis) and −log10p (Y-axis) for the 730 proteins tested for breast cancer risk in the Mendelian randomisation analysis. (Mälarstig, A. et al., 2023)
- Cardiometabolic Risk Stratification: Using the Olink Explore 3072 PEA platform, researchers analyzed plasma proteomic profiles from 53,030 UK Biobank participants, assessing 2,923 proteins. A protein-wide association study (PWAS) investigated delta-BMI associations, adjusted for covariates like BMI, sex, and age. The PWAS approach, visualized through Manhattan plots, highlighted robust protein-disease relationships. This underscores Olink's role in uncovering biological pathways for cardiometabolic risk stratification.
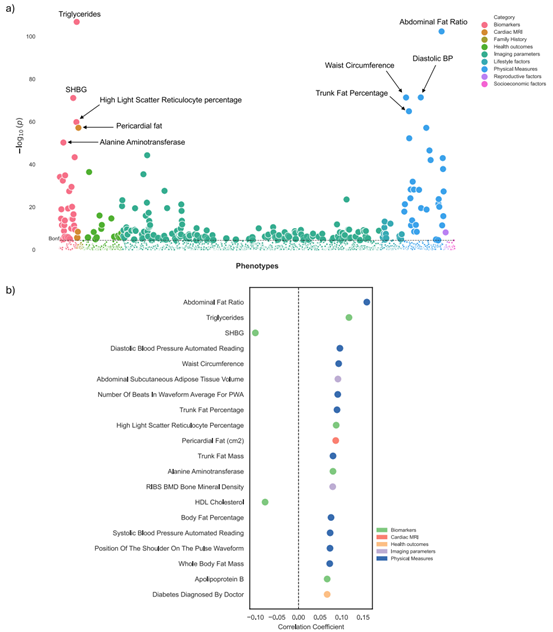 Figure 5: Phenome-wide association study (PheWAS) of delta-BMI in the BIDMC cohort. (Pastika, L. et al., 2024)
Figure 5: Phenome-wide association study (PheWAS) of delta-BMI in the BIDMC cohort. (Pastika, L. et al., 2024)
Conclusion: Transforming Biomedical Discovery Through Advanced Proteomics
A Paradigm Shift in Protein Analytics
The Olink Explore 3072 platform represents a fundamental transformation in large-scale protein analysis, overcoming long-standing limitations that have hindered proteomic research. By combining exceptional analytical specificity with unprecedented multiplexing capabilities, this platform enables investigations previously constrained by technical feasibility.
Innovative Architecture for Comprehensive Proteomics
The platform's distinctive architecture partitions 3,072 protein targets across eight specialized panels, creating an optimal balance between comprehensiveness and analytical performance. This design is complemented by rigorous quality control procedures that ensure data reliability across diverse sample types and study designs.
Key architectural advantages:
- Specialized panels enable targeted protein profiling across biological domains.
- Modular design allows researchers to select relevant panels for their specific research questions.
- Standardized procedures ensure consistency across experiments and research groups.
Broad Applications Across Research Domains
The protein measurements generated by the platform have demonstrated remarkable utility across diverse research domains:
- Neurological degeneration: Revealing protein signatures associated with disease progression.
- Cancer biology: Identifying biomarkers for early detection and therapeutic targeting.
- Metabolic adaptation: Uncovering systemic responses to physiological challenges.
Evolution and Future Directions
As proteomic technologies continue to evolve, the Olink platform remains at the forefront of biomedical discovery. This is particularly relevant as large-scale biobanks increasingly incorporate protein biomarkers alongside genomic and clinical data.
The recent expansion to 5,400-plex analysis (Explore HT) significantly extends the discovery potential while maintaining backward compatibility with existing datasets. This ensures that previous research investments remain valuable while accessing enhanced capabilities.
Key Benefits for Researchers
For academic and biotechnology researchers navigating the complex landscape of proteomic technologies, the Olink Explore 3072 platform offers a compelling combination of:
- Analytical performance: High specificity and sensitivity for reliable results.
- Biological coverage: Comprehensive profiling across multiple protein classes.
- Computational robustness: Data compatibility with advanced analysis methods.
FAQs — Olink vs SomaScan
1. What are the specialized panels, and can they be used flexibly?
The platform's comprehensive coverage is achieved through a modular design consisting of eight panels, each focusing on specific biological domains, such as Cardiometabolic, Inflammation, Neurology, and Oncology. These panels can indeed be mixed and matched based on research objectives, allowing for a highly flexible and cost-effective experimental design.
2. What types of samples are compatible with the platform?
The platform accommodates a wide range of sample types, including serum, plasma, and cerebrospinal fluid (CSF).
3. What is the data output format, and how is quality control ensured?
The primary data output is Normalized Protein eXpression (NPX) values, which are relative quantitative units. The workflow includes a robust quality control (QC) steps. After the immuno-reaction and amplification, libraries undergo quality assessment, for example, via an electropherogram to confirm the expected amplicon size, before sequencing. Data is also processed to flag assays or samples with potential QC issues.
4. Can data from Olink Explore 3072 be integrated with newer Olink products?
Yes, Olink provides bridging protocols to normalize and combine datasets from different product generations. Specifically, tools exist to bridge data from the Explore 3072 platform to the newer Explore HT (5,400-plex) or Olink Reveal products, facilitating meta-analyses and longitudinal studies.
Related Reading
To better understand how Olink performs against other proteomic platforms, we recommend exploring the following related guides:
Understanding Olink's Data Analysis Process: From Raw Data to Insights — An overview of how Olink's structured analytical pipeline transforms raw protein data into biological insights through quality control, differential analysis, and functional interpretation.
Olink Proteomics for Large-Scale Population Studies: A Success Story — An overview of how Olink's proteomics platform is advancing large-scale population studies through high-quality biomarker discovery and causal insights.
Olink Explore 3072 vs Explore 1536: Selecting the Right Platform for Your Study Scale — A practical guide to selecting the right throughput platform to maximize efficiency and power in your proteomic study.
References
- Dovgan, I., Koniev, O., Kolodych, S. et al. Antibody–Oligonucleotide Conjugates as Therapeutic, Imaging, and Detection Agents. Bioconjug. Chem. 30, 2483-2501 (2019).
- Michael P.A. D., Takahiro S., Haitham A. et al.Plasma protein biomarkers for early prediction of lung cancer. eBioMedicine 93,104686 (2023).
- Chia, R., Moaddel, R., Kwan, J.Y. et al. A plasma proteomics-based candidate biomarker panel predictive of amyotrophic lateral sclerosis. Nat Med 31, 3440–3450 (2025).
- Mälarstig, A., Grassmann, F., Dahl, L. et al. Evaluation of circulating plasma proteins in breast cancer using Mendelian randomisation. Nat Commun 14, 7680 (2023).
- Pastika, L., Sau, A., Patlatzoglou, K. et al. Artificial intelligence-enhanced electrocardiography derived body mass index as a predictor of future cardiometabolic disease. npj Digit. Med. 7, 167 (2024).


