Introduction
Olink's Target 48 immune panels give you focused, high-sensitivity coverage of cytokines and chemokines central to T helper (Th) biology and leukocyte trafficking—ideal for translational studies where sample volume is tight and mechanistic clarity matters. In practical terms, "Target 48 Th1/Th2/Th17 and chemokine coverage" means you can quantify key mediators across Th1, Th2, Th17, and Treg contexts while tracking chemokine signals that move cells between lymphoid tissues and inflamed sites.
Why does a 48-plex format help? Because it concentrates on the most decision-relevant proteins and runs on ~1 µL input per panel, enabling dense longitudinal sampling, pediatric or rare-specimen cohorts, and multi-center designs without overtaxing biobanks. And by reporting in both absolute pg/mL (for Target 48 Cytokine) and NPX (a normalized, relative log2 unit used across Olink assays), you can support discovery, cross-plate comparability, and downstream integration with other omics.
Key takeaways
- ~1 µL sample input supports longitudinal, pediatric, and scarce-sample designs.
- Balanced coverage across Th1/Th2/Th17/Treg and chemokines enables pathway-level interpretation.
- Target 48 Cytokine provides absolute pg/mL alongside NPX, improving comparability within validated ranges.
- Robust QC, LOD/LOQ handling, and inter-plate calibration are essential for multi-plate or multi-center studies.
Coverage overview
The Target 48 family emphasizes immune mediators relevant to Th differentiation, effector function, and cell trafficking. Exact analyte lists can vary by panel version; always confirm the assay roster and validation files before a study. Below is an axis-based framing to guide interpretation and study design.
Olink publishes the canonical panel roster and per‑analyte validation in the Target 48 panel content and kit documents; the Cytokine panel typically lists ~45 proteins (plus controls). For exact analyte lists and limits of detection/quantification, check Creative Proteomics' Target 48 service page.
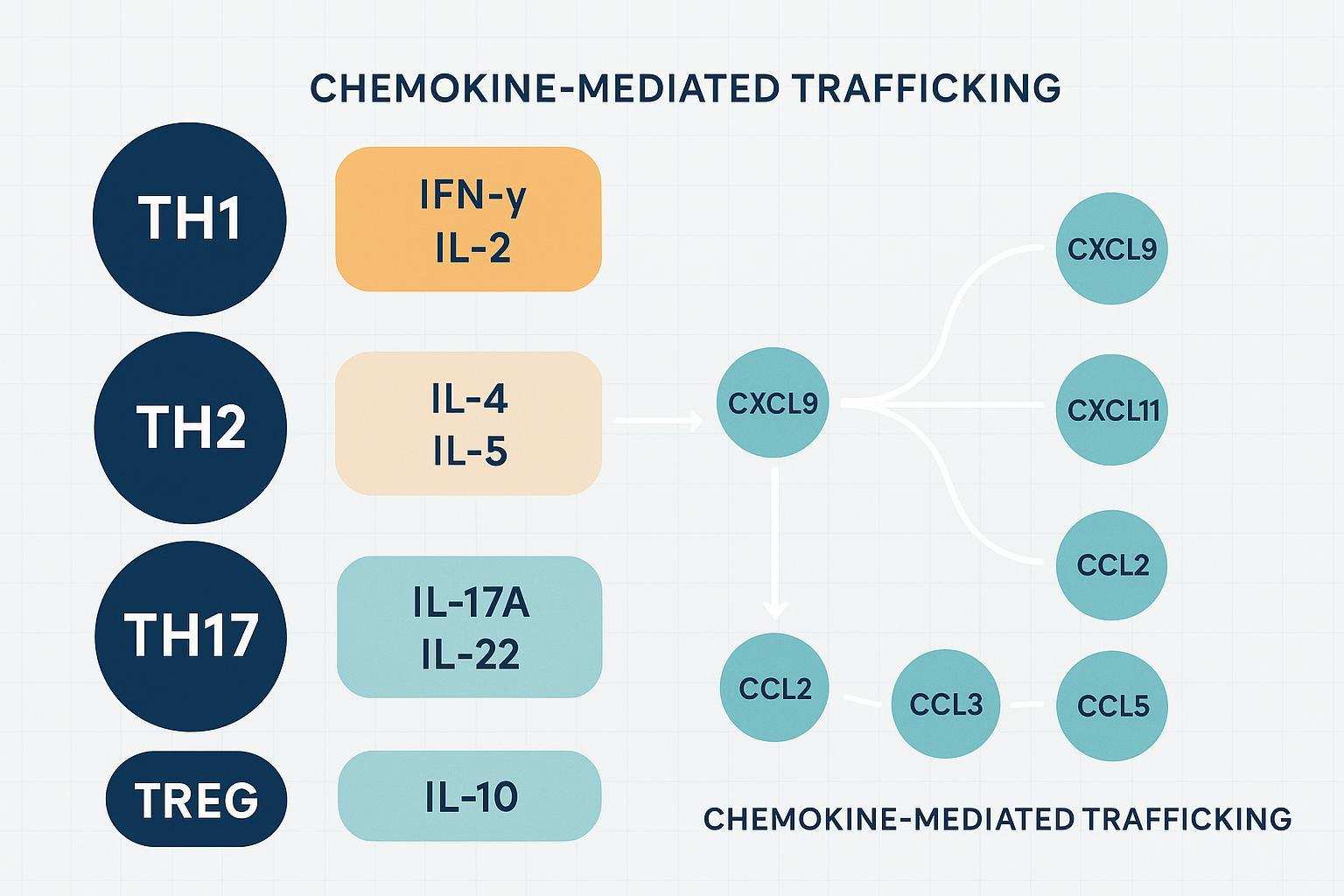
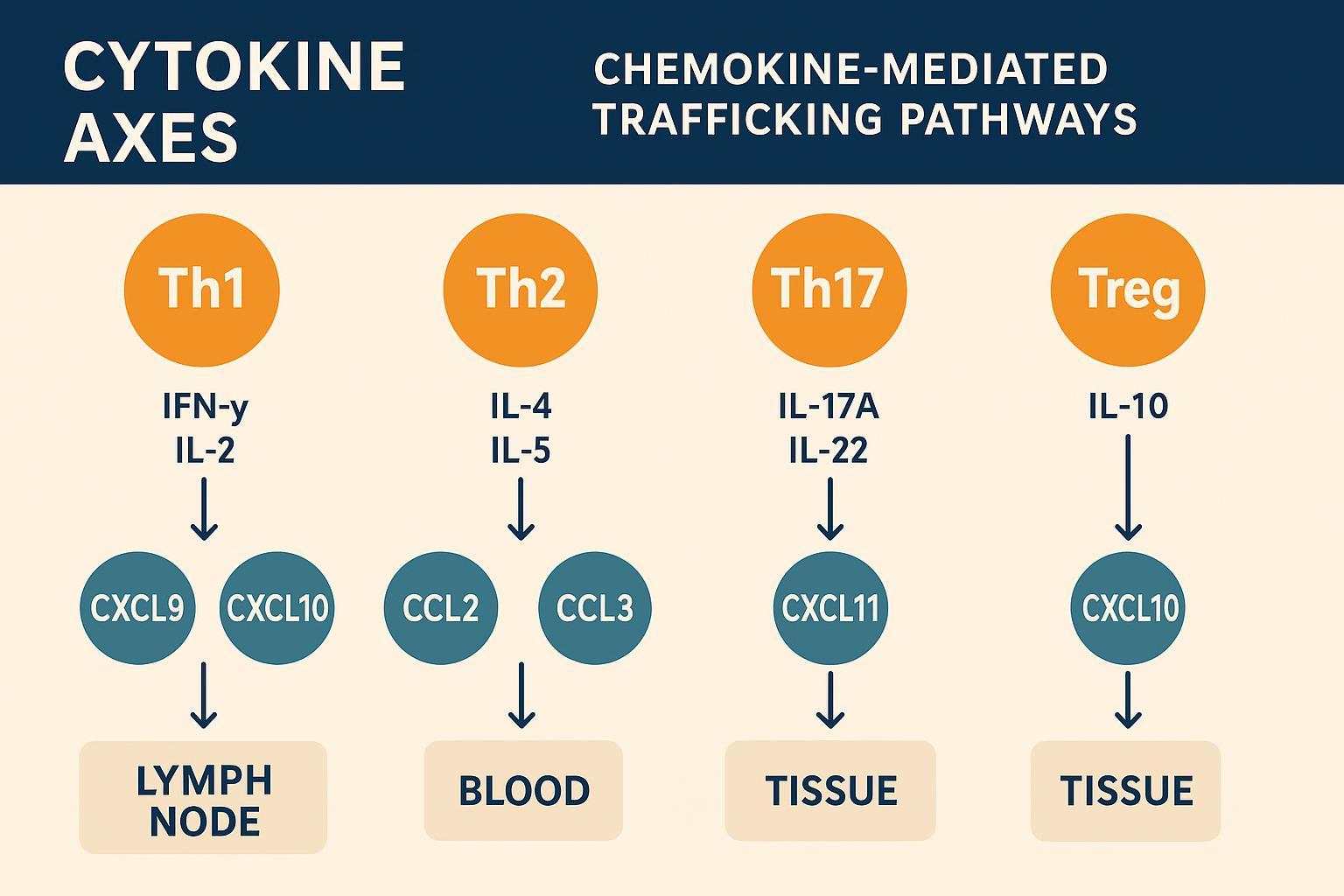
Th1 markers and readouts
Th1 responses often center on IFN-γ (IFNG) and IL-2 (IL2), with downstream chemokines such as CXCL9, CXCL10, and CXCL11 that recruit CXCR3+ T cells to inflamed tissues. In vaccine and infection contexts, rises in IFN-γ–inducible chemokines are common correlates of cell-mediated immunity. This is where Olink Target 48 Th1/Th2/Th17 and chemokine coverage helps you connect cytokine surges with trafficking signals to interpret immune kinetics.
Th2/Th17 axes and Treg context
Th2 signaling (e.g., IL‑4, IL‑5) relates to humoral responses and allergic inflammation, while Th17 cytokines (e.g., IL‑17A, IL‑22) link to neutrophil recruitment and barrier immunity. A Treg‑associated milieu often includes IL‑10 and other anti‑inflammatory cues. In autoimmunity, shifts in the Th17/Treg balance can flag disease activity; in oncology, Th1 vs. Th2 skew can reflect tumor microenvironment dynamics.
Chemokines and trafficking signals
Chemokines orchestrate movement: IFN‑γ‑inducible CXCL9/10/11 draw T cells; CCL2/3/5 shape monocyte and T‑cell homing. Tracking these signals alongside cytokines helps explain why cells appear where they do—and when. In studies of infection or immunotherapy, changes in chemokine gradients often mirror functional shifts in immune infiltration.
Technical specifications
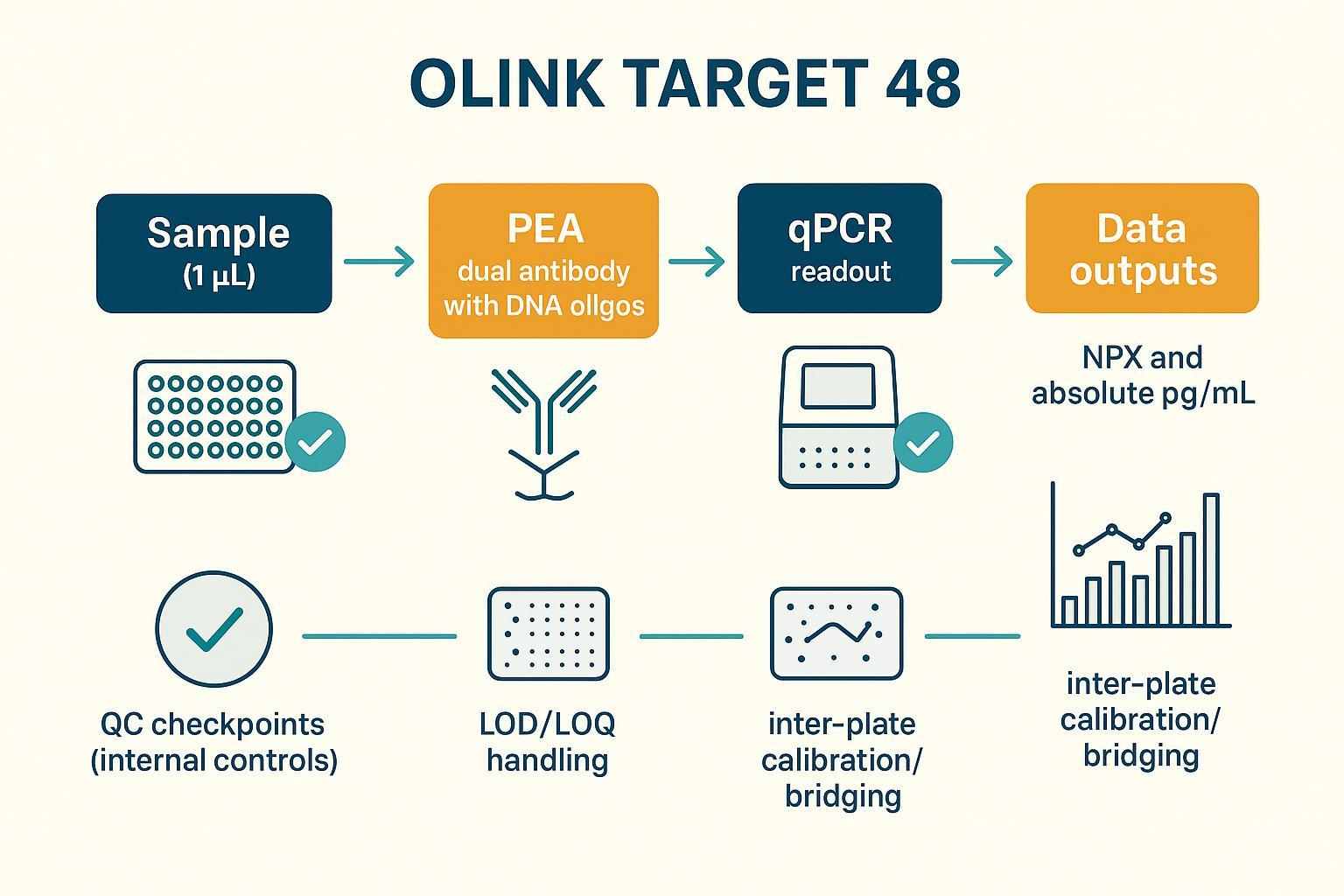
PEA principle, qPCR readout, and 1 µL input
Olink's Proximity Extension Assay (PEA) uses dual antibodies carrying DNA oligonucleotides that bind the same protein. When both bind, their oligos come into proximity, are extended to form a unique DNA tag, and then quantified by qPCR. This chemistry enables multiplex detection from approximately 1 µL per sample, supporting studies where volume constraints are real. See Olink's overview of the technology in the official description of what PEA is and instrument details on the Signature Q100 page .
Reporting formats: pg/mL and NPX normalization
- NPX (Normalized Protein eXpression) is a relative, log2‑scaled unit derived from qPCR Cq/Ct values through assay‑specific normalization. It's ideal for within‑study comparisons and signature building; equal NPX across different proteins doesn't imply equal concentration. Olink clarifies the role of NPX and normalization in their FAQ .
- Absolute pg/mL is available by default on Target 48 Cytokine, using calibrators to determine concentrations within validated ranges. This supports cross‑study comparability and practical thresholds, provided values stay within LOQ/ULOQ. Details are outlined in the Target 48 brochure and specific assay pages like interleukin‑2 .
- Interpretation nuance: Use NPX for robust pattern detection and modeling, and pg/mL when you need concentration values—for example, correlating with clinical labs or setting cut‑offs. Avoid back‑calculating NPX to pg/mL outside validated workflows.
QC, LOD handling, and inter-plate calibration
A rigorous QC framework is built into Olink's workflow, monitoring incubation, extension, and detection phases with internal/external controls. Assay‑specific LOD/LOQ define the measurement range; values below LOD should be censored or handled consistently (e.g., set to LOD/√2 or use model‑based approaches), and values near LOQ require caution. For multi‑plate studies, include overlapping bridge samples and apply plate centering or distribution alignment. Olink's recommended strategies appear in their stepwise guide to multiplex immunoassay analysis and the OlinkAnalyze normalization documentation .
Applications and study design
Use cases: autoimmunity, infection, oncology, vaccines
- Autoimmunity: Track Th17‑linked inflammation (e.g., IL‑17 family contexts), Th1 chemokines (CXCL9/10/11), and anti‑inflammatory signals (IL‑10). Longitudinal sampling helps distinguish flare vs. remission trends; replicate design and pre‑defined LOD handling reduce bias.
- Infection and vaccines: Monitor Th1/Th2 balance and chemokine‑mediated trafficking during acute and memory phases. The ~1 µL input allows dense time‑courses across visits; batching by visit reduces confounding, while bridge samples ensure inter‑plate alignment.
- Oncology: Profile IFN‑γ‑inducible chemokines, Th skew, and immuno‑regulatory cytokines to infer tumor microenvironment activity. Pair proteomic readouts with transcriptomic signatures or orthogonal assays for validation.
Authoritative references and reporting standards: For translational contexts, see Nature Communications 2021 severe COVID‑19 plasma proteomics using Olink PEA, which demonstrates 1 µL plasma workflows and cytokine/chemokine profiling in hospitalized cohorts, and Wik et al.'s 2021 validation comparing Olink Target 48 Cytokine, detailing qPCR readout performance across cytokines. For community reporting norms, align study metadata and QC with the HUPO‑PSI standards portal (MIAPE and related specifications), noting module‑based guidance for reproducibility.
Panel selection and adjacent options
Combine Target 48 Cytokine, which offers absolute pg/mL reporting, with Immune Surveillance to broaden coverage across Th axes and chemokine pathways. For hypothesis‑driven projects, custom Flex configurations can prioritize specific mediators. For mouse models, the dedicated preclinical Target 48 Mouse Cytokine option extends similar logic to murine studies. Always confirm assay lists and validation ranges before finalizing the design.
Sample handling, batching, and analysis tips
- Pre‑analytics: Aliquot promptly, avoid freeze–thaw cycles, and store at −80 °C. Randomize plate positions to minimize positional effects.
- Batching: Group samples by visit or clinical subgroup; include 8–12 bridge samples across plates to enable normalization.
- Analysis: Inspect QC flags; decide on <LOD handling upfront; report NPX for discovery and pg/mL where available for comparability. For deeper method context, see the Creative Proteomics knowledge pages on PEA technology and Olink data analysis and normalization .
Disclosure: Creative Proteomics is our product.
A neutral micro‑case: In a vaccine cohort, a longitudinal Target 48 Cytokine study tracked Th1/Th2 balance and IFN‑γ‑linked chemokines across five timepoints using ~1 µL per draw. Bridge samples aligned data across plates; NPX trends highlighted peak response windows, while pg/mL supported cross‑study comparisons within LOQ. Downstream PBMC RNA‑seq and targeted metabolomics assessed transcriptional and metabolic correlates of cytokine changes, strengthening mechanistic interpretation. For end‑to‑end Olink Target 48 services and integration options, see the Target 48 Cytokine Panel overview and multi‑omics integration.
Conclusion
Focused 48‑plex coverage across Th1/Th2/Th17/Treg axes and chemokines gives translational teams the right balance of mechanism and practicality—especially when ~1 µL input makes dense sampling feasible. With NPX for robust pattern detection and pg/mL for validated comparability, plus solid QC and normalization, Target 48 panels fit autoimmunity, infection/vaccine, and oncology studies.
Practical next steps: Define your biological questions by axis (Th1, Th2/Th17/Treg, chemokines), confirm the exact assay list and LOQ/LOD per marker, plan plate layouts with bridge samples, and pre‑register LOD handling rules. If you need support from study design through multi‑omics follow‑up, explore neutral, end‑to‑end Olink services and integration pathways via Creative Proteomics' resources.
Frequently asked questions (FAQ)
Q1 — What is the practical difference between NPX and pg/mL for Target 48 data?
NPX (Normalized Protein eXpression) is a relative, log2-scaled unit produced after assay-specific normalization; it is optimized for within-study comparisons and signature discovery. Absolute pg/mL values from Target 48 Cytokine use calibrators and support cross-study comparability within each assay's validated LOQ/ULOQ. For technical details on NPX and calibration, see Olink's FAQ on NPX and data units and the Target 48 brochure on absolute quantification.
Q2 — Can I convert NPX to pg/mL directly?
No. NPX and pg/mL are produced through different processing steps; NPX is a normalized qPCR-derived relative unit, while pg/mL requires a calibrated standard curve. Converting NPX to pg/mL outside validated assay workflows is not advised. If absolute concentrations are required, use the panel's calibrated pg/mL outputs where available and follow assay-specific LOQ guidance in the Target 48 user documentation.
Q3 — Is 1 µL input reliable for Target 48 runs and what are common caveats?
Target 48 formats are validated to work with approximately 1 µL of plasma or serum, enabling dense longitudinal or low-volume studies. Common caveats: matrix effects, hemolysis, or low total protein can affect assay performance; always follow pre-analytic SOPs (aliquoting, avoiding freeze–thaw) and inspect QC flags. Instrument context (e.g., Signature Q100) and sample-matrix notes are documented by Olink and should be reviewed before study start: see the Signature Q100 information.
Q4 — How should values below LOD or near LOQ be handled in analysis?
Handle <LOD and near‑LOQ values consistently and document the rule a priori. Common approaches include censoring (report as <LOD), substitution (LOD/2 or LOD/√2) when justified, or model-based methods that treat left-censoring explicitly. Which approach to choose depends on the fraction of censored values and downstream analysis; Olink's validation materials and community resources (e.g., normalization and QC guides) provide context for selecting an approach: see Olink's stepwise guide to multiplex immunoassay analysis.
Q5 — What is the recommended practice for inter‑plate normalization or bridging across runs?
Include overlapping bridge samples across plates (biological replicates or pooled controls) and apply plate-centering or distribution alignment methods to remove technical offsets. Document the bridging design in the study plan, visualize pre- and post-normalization distributions, and consider established tools such as the OlinkAnalyze normalization documentation for method options.
Q6 — How do I choose between Target 48 panels or Flex/custom options for a specific study question?
Choose a Target 48 panel when you need a focused, validated set of cytokines/chemokines with low-volume requirements and, where available, calibrated pg/mL outputs. Use Flex/custom selections if your hypothesis requires non‑standard analytes or a specific subset not covered by off‑the‑shelf panels. Confirm analyte lists and LOQ/ULOQ for candidate panels before ordering to ensure critical markers are included.
Q7 — How can proteomic results be integrated with other omics for mechanistic insight?
Integrate Target 48 protein readouts with transcriptomics, metabolomics, or targeted mass spectrometry to triangulate pathway signals and strengthen mechanistic interpretation. Practical workflows include correlating temporal NPX trajectories with gene expression changes in matched PBMC RNA‑seq or using targeted metabolomics to probe energy and inflammatory metabolites that co-vary with cytokine peaks. For a service-oriented example of end‑to‑end Target 48 execution plus multi‑omics integration, see Creative Proteomics' Olink multi‑omics integration overview.
Creative Proteomics — Multi‑Omics Research Partner & Olink Proteomics Services Provider. Creative Proteomics supports discovery‑to‑preclinical studies with Olink PEA‑based proteomics and integrated multi‑omics workflows, combining experienced laboratory teams, standardized QC, and translational study design support. For questions about study design or data interpretation, contact the team via /about.



