Why Proper Olink Explore 3072 Implementation Matters for Biobank Science
Large-scale proteomic studies in biobank settings present unique challenges: sample volume limitations, batch effects across thousands of samples, and the need for reproducible, high-quality data. Implementing Olink Explore 3072 with proper infrastructure and standardized protocols addresses these challenges by enabling robust protein quantification while minimizing technical variability. This guide provides biobank teams with a comprehensive framework for establishing Olink workflows that yield publication-ready proteomic data.
What Is Olink Explore 3072 and Core Advantages for Population Studies
Olink Explore 3072 utilizes Proximity Extension Assay (PEA) technology to simultaneously quantify 2,941 proteins from minute sample volumes (1-3 μL). This platform offers distinct advantages for biobank-scale research:
Key Advantages for Large Cohorts:
| Advantage | Practical Benefit for Biobanks |
| High multiplexing capacity | Measure thousands of proteins from precious biobank samples without volume depletion |
| Exceptional specificity | Dual antibody recognition minimizes false positives in large-scale screening |
| Low sample volume requirements | Ideal for longitudinal studies or multi-omics integration where sample availability is limited |
| High reproducibility | Median CV <10% ensures data consistency across batches and timepoints |
| Scalable throughput | Process hundreds to thousands of samples with standardized protocols |
Comparative Performance Context
- Versus mass spectrometry: Better sensitivity for low-abundance proteins without need for sample prefractionation.
- Versus traditional immunoassays: Superior multiplexing capacity and specificity through DNA-conjugated antibody technology.
- For genetic association studies: Higher concordance with genetic data (cis-pQTL detection) strengthens proteogenomic analyses.
Infrastructure & Laboratory Requirements for Biobank-Scale Olink Operations
Establishing Olink Explore 3072 workflows requires specific infrastructure considerations for large-scale operations:
1. Essential Laboratory Infrastructure
- Dedicated pre-PCR and post-PCR areas to prevent amplicon contamination across high-throughput processing
- Temperature-stable workspaces with minimal vibration for consistent assay performance
- -80°C archival storage with monitoring systems for long-term sample integrity
- Refrigerated centrifuges (4°C) with balanced rotors for standardized sample processing
2. Critical Equipment Specifications
| Function | Equipment Requirements | Specifications for Large Studies |
| Sample processing | Refrigerated centrifuges | Capacity for 96-well plates, 2,000-2,500 × g capability |
| Liquid handling | Automated liquid handlers | Low-volume precision (1-10 μL), 96- or 384-well capability |
| Incubation | Thermal mixers/hybridizers | Stable temperature control (4°C-37°C), capacity for full plates |
| Amplification & sequencing | Illumina sequencers | NovaSeq 6000/NextSeq 1000-2000 for high-throughput processing |
| Data management | High-performance computing | Storage and processing capacity for thousands of samples |
Sample Collection, Handling and Pre-Analytical Considerations for Biobanks
Standardized Sample Collection Protocols
- Preferred sample type: EDTA plasma demonstrates superior performance characteristics
- Collection tubes: Standardized across collection sites to minimize pre-analytical variability
- Processing timeline: Centrifugation within 4 hours of collection at 2,000 × g for 10 minutes (4°C)
- Aliquoting strategy: Single-use aliquots to minimize freeze-thaw cycles
Pre-Analytical Quality Control Measures
- Hemolysis index documentation for each sample
- Lipemia assessment to identify potentially problematic samples
- Aliquot volume standardization (≥40 μL recommended for repeat analyses)
- Comprehensive sample metadata tracking including processing timelines
Protocol Workflow: Optimized for Large-Scale Biobank Operations
Step 1: Sample Plate Preparation and Normalization
- Thaw samples on ice or at 4°C with minimal freeze-thaw cycles
- Dilute samples according to panel-specific requirements
- Implement block randomization across plates to balance case/control status
Step 2: PEA Incubation and Extension
- Incubate samples with Olink probe mix (overnight at 4°C standard)
- Perform proximity extension with DNA polymerase
- Include internal controls in each sample for quality monitoring
Step 3: Amplification and Library Preparation
- Pool samples following Olink Explore 3072 specifications
- Prepare sequencing libraries with dual indexing
- Implement bridge samples across plates for batch effect correction
Step 4: Sequencing and Data Generation
- Sequence on Illumina platforms following Olink-recommended parameters
- Include PhiX spike-in for run quality monitoring
- Generate FASTQ files for downstream processing
Quality Control Framework for Large-Scale Studies
Multi-Level QC Protocol
| QC Level | Assessment Metrics | Acceptance Criteria |
| Sample-level QC | Detection rate, internal controls | >80% proteins detected, control deviation <0.3 NPX |
| Plate-level QC | Plate controls, sample correlations | CV <10% for controls, high replicate correlation |
| Study-level QC | Batch effects, population outliers | PCA clustering by biology not batch, <5% sample exclusion |
Batch Effect Mitigation Strategy
- Reference samples: Include pooled plasma controls on each plate
- Balanced plate design: Distribute biological groups evenly across plates
- Staggered processing: Process case/control samples simultaneously across batches
- Statistical correction: Implement ComBat or similar methods for batch adjustment
Cost, Throughput and Scaling Considerations for Biobanks
Economic Modeling for Large Cohorts
- Per-sample cost optimization through batch size maximization
- Reagent procurement strategy for consistent lot usage across large studies
- Sequencing capacity planning to maintain project timelines
Throughput Optimization Framework
- Plate-based processing enables 96-384 samples per batch
- Workflow parallelization for simultaneous sample processing
- Automated liquid handling implementation reduces hands-on time
Troubleshooting Common Large-Scale Implementation Challenges
Problem: High Inter-Plate Variability
- Solution: Implement robust bridge sample normalization, standardize reagent lots
Problem: Sample Detection Rate Issues
- Solution: Verify sample quality metrics, optimize dilution factors, check pipette calibration
Problem: Batch Effects in Data Analysis
- Solution: Enhance balanced study design, include technical covariates in statistical models
Case Studies
The following case studies highlight how the Olink Explore platform is being successfully implemented in large-scale population studies, demonstrating its utility in generating biologically actionable insights.
- Case Study 1
This study by Shah et al. explores large-scale plasma proteomics to identify novel proteins and networks linked to heart failure (HF) development. The research leveraged the SomaScan v4 aptamer-based assay to measure 4,877 plasma proteins in 13,900 HF-free individuals across diverse cohorts, revealing 37 proteins consistently associated with incident HF. A critical component of this work was the orthogonal validation of candidate proteins using the Olink Explore 3072 proximity extension assay, which confirmed the specificity of 27 out of 37 HF-associated proteins. The integration of Olink 3072 enhanced the reliability of proteomic findings, supporting Mendelian randomization analyses that implicated causal pathways and druggable targets, thereby bridging genomic insights with HF pathophysiology.
 Figure 1: Schematic overview of study design. (Shah, A.M. et al., 2024)
Figure 1: Schematic overview of study design. (Shah, A.M. et al., 2024)
- Case Study 2
In the UK Biobank Pharma Proteomics Project by Sun et al., plasma proteomic profiles of 54,219 participants were characterized to link genetics with health outcomes. The study predominantly utilized the Olink Explore 3072 platform, an antibody-based proximity extension assay, to measure 2,923 unique proteins, enabling high-throughput pQTL mapping and association analyses with diseases. The Olink 3072 technology was instrumental in achieving robust data quality, with low batch effects and high reproducibility, leading to the discovery of 14,287 primary genetic associations and insights into trans-pQTL networks. This resource underscores Olink 3072's role in advancing drug discovery by providing a deep proteogenomic atlas for the scientific community.
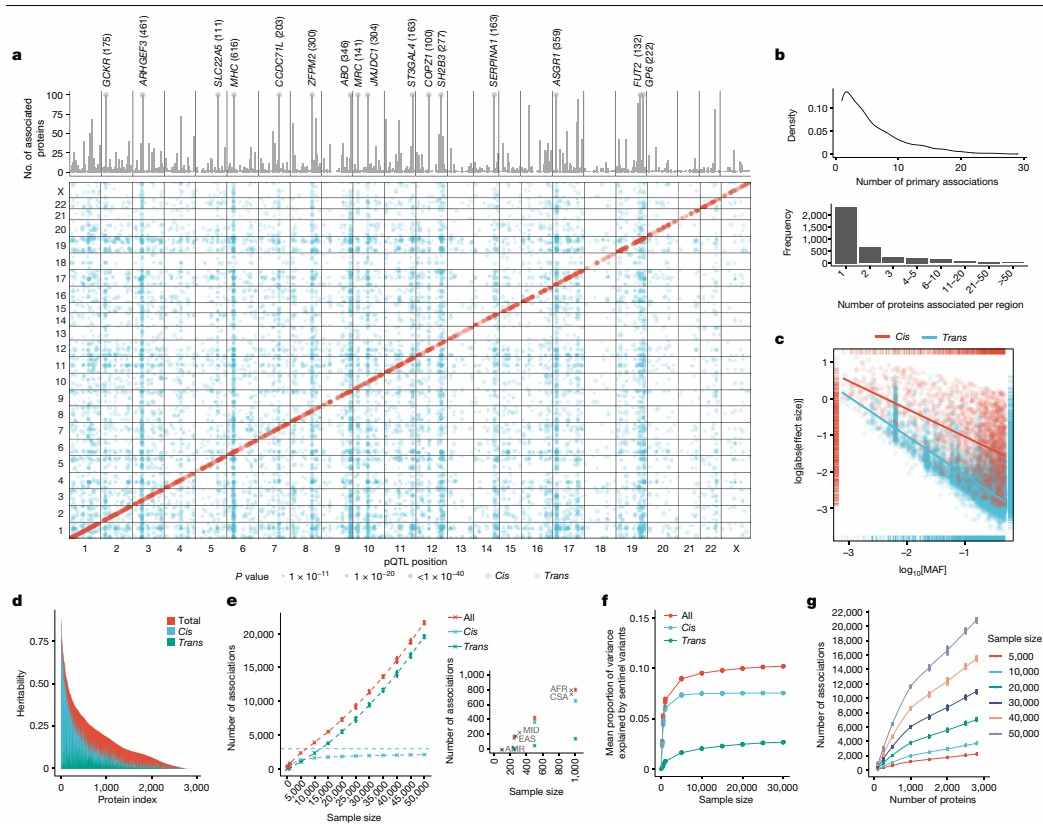 Figure 2: The genetic architecture of pQTLs. (Sun, B.B. et al., 2023)
Figure 2: The genetic architecture of pQTLs. (Sun, B.B. et al., 2023)
- Case Study 3
In this comprehensive comparative study published in Communications Chemistry, researchers conducted a detailed evaluation of the Olink Explore 3072 platform alongside a peptide fractionation-based mass spectrometry (HiRIEF LC-MS/MS) method for plasma proteomics profiling across 88 samples. The study demonstrated that Olink Explore 3072 played a critical role in enabling robust, multiplexed quantification of 2,913 proteins with high precision (median technical CV of 6.3%) and superior sensitivity for low-abundance proteins, complementing MS-based methods which excelled in mid-to-high abundance protein detection.
Notably, Olink Explore 3072 exhibited strong quantitative agreement with MS (median Spearman correlation ρ=0.59), facilitated the identification of sex-specific protein differences with high concordance, and supported the development of the PeptAffinity tool for peptide-level cross-platform validation, underscoring its utility in large-scale biomarker studies where throughput and reliability are paramount.
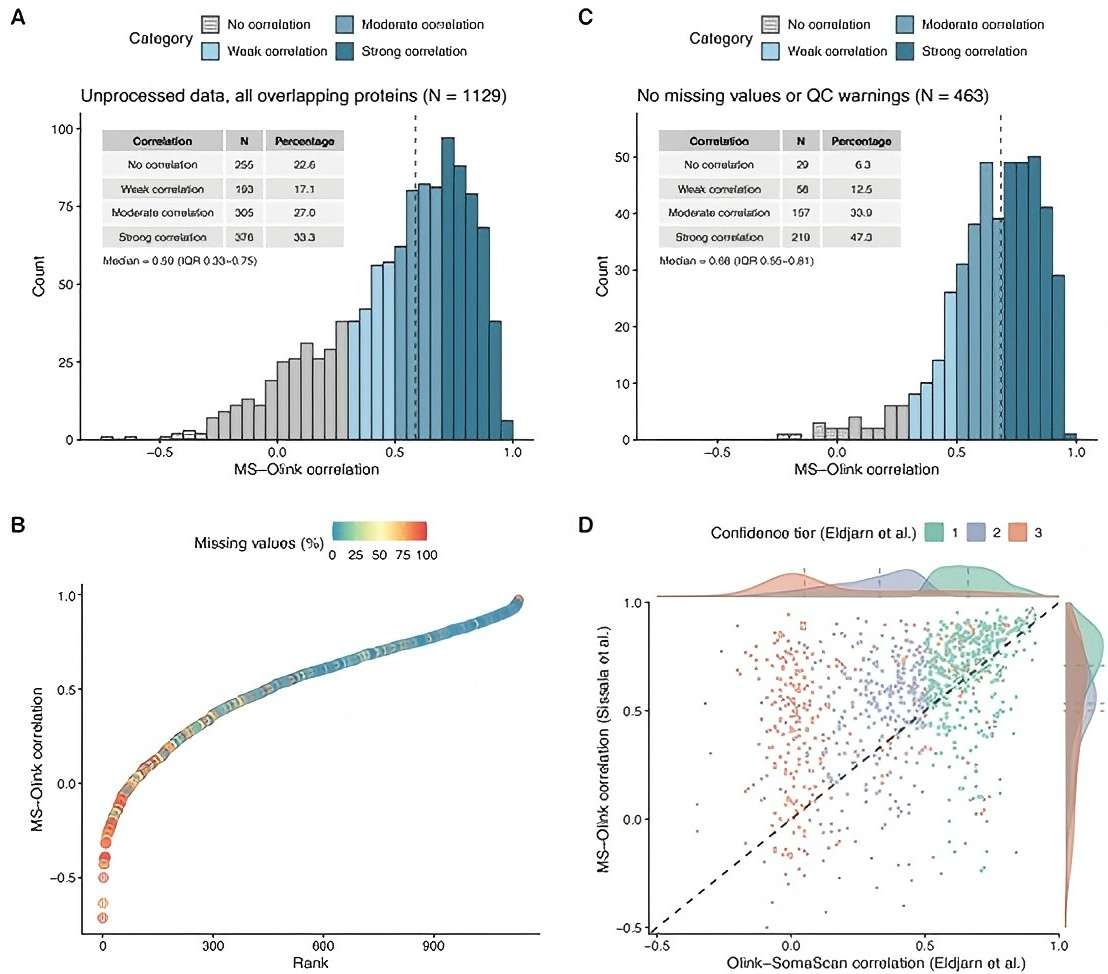 Figure 3: Cross-platform correlation of protein levels. (Sissala, N. et al., 2025)
Figure 3: Cross-platform correlation of protein levels. (Sissala, N. et al., 2025)
Conclusion: Implementing Robust Olink Explore 3072 Workflows in Biobank Settings
Establishing standardized Olink Explore 3072 protocols in biobank environments enables high-quality proteomic profiling at population scale. Through careful attention to infrastructure requirements, pre-analytical standardization, and rigorous quality control, research teams can generate proteomic data suitable for genetic association studies, biomarker discovery, and multi-omics integration.
The framework presented here provides biobank scientists with a comprehensive roadmap for implementing Olink technologies that yield reproducible, high-fidelity proteomic data capable of powering large-scale biomedical discoveries.
FAQs
1. How should samples be collected and processed to ensure data quality?
- Standardized Protocols: Use consistent blood collection tubes (e.g., EDTA plasma) and processing timelines (centrifuge within 4 hours of collection).
- Minimize Pre-Analytical Variability: Avoid repeated freeze-thaw cycles; aliquot samples for single-use.
- Quality Controls: Include inter-plate controls (e.g., pooled reference samples) to monitor batch effects.
- Document Metadata: Track hemolysis, lipemia, and processing delays to identify confounders.
2. What study design strategies mitigate batch effects?
- Randomization: Distribute samples from different groups (e.g., case/control) evenly across plates and processing batches.
- Bridge Samples: Incorporate shared reference samples in each batch to enable cross-run normalization.
- Balanced Processing: Ensure samples from all experimental groups are processed simultaneously to avoid time-dependent biases.
3. How can researchers address missing data in Olink datasets?
- Prevention: Ensure sufficient sample volume and avoid technical artifacts (e.g., pipetting errors).
- Imputation: Use methods like minimum value imputation for values below LOD, but document assumptions.
- Statistical Adjustments: In differential analysis, apply models that account for missingness (e.g., mixed-effects models).
4. How do I interpret and validate protein-disease associations?
- Mendelian Randomization (MR): Use genetic instruments (pQTLs) to infer causal relationships between proteins and diseases.
- Replication: Validate findings in independent cohorts or public datasets (e.g., UK Biobank, HUNT).
- Biological Context: Integrate pathway analyses (e.g., GO, KEGG) to prioritize mechanistically plausible biomarkers.
Related Reading
For deeper insights, we recommend exploring the following related guides:
Olink Proteomics for Large-Scale Population Studies: A Success Story — A Success Story of how large-scale plasma proteomics accelerates biobank research, enables long-term risk prediction for diverse diseases, and reveals novel disease mechanisms and therapeutic targets through highly multiplexed and sensitive assays.
A Deep Dive into the Olink Explore 3072 Platform: Expanding the Boundaries of Proteomic Discovery — A vision of how Olink's scalable, high-throughput proteomics is accelerating biomarker discovery, reshaping drug development, and unlocking the functional proteome through ultra-sensitive, large-scale studies of complex biological samples.
References
- Shah, A.M., Myhre, P.L., Arthur, V. et al. Large scale plasma proteomics identifies novel proteins and protein networks associated with heart failure development. Nat Commun 15, 528 (2024).
- Sun, B.B., Chiou, J., Traylor, M. et al. Plasma proteomic associations with genetics and health in the UK Biobank. Nature 622, 329–338 (2023).
- Sissala, N., Babačić, H., Leo, I.R. et al. Comparative evaluation of Olink Explore 3072 and mass spectrometry with peptide fractionation for plasma proteomics. Commun Chem 8, 327 (2025).







