Large clinical cohorts are now the norm: hundreds to over a thousand samples per study, multi-center sourcing, tight timelines, and high compliance expectations. The pivotal question is no longer whether proteomics is insightful—it's whether you can deliver results at scale with reproducibility and an audit trail. This ultimate guide focuses on the execution layer of Olink-based proteomics: how to scale without sacrificing data quality, how to design batches and bridge samples, and how to make immune profiling feasible in clinical contexts.
Key takeaways
- Large-scale Olink proteomics is feasible when you treat scalability and QC as first-class design constraints—bridge samples, cross-batch alignment, and NPX normalization are non-negotiable.
- Precision and detectability vary by analyte and matrix; plan pilots to establish CV ranges and LOD stability before full rollout.
- Clinical cohort immune profiling is realistic but demands careful handling of cytokine-poor contexts and below-LOD data.
- Pre-analytical harmonization across sites (collection, storage, shipping) drives success as much as the platform does.
- Integrate proteomics with multi-omics early—pQTL/proteogenomics and orthogonal validation help turn exploratory signals into decision-grade biomarkers.
What "high-throughput" really means for Olink at cohort scale
"High-throughput" in Olink-based cohorts spans four dimensions: sample processing capacity, protein coverage, data consistency across time, and operational robustness. Proximity Extension Assay (PEA) is well-suited for cohorts because it separates affinity binding from NGS readout, enabling minimal sample input (often 1–6 µL) and strong specificity with negligible cross-reactivity.
- Throughput and coverage: Explore HT measures 5,400+ proteins with NGS readout and is engineered for high sample throughput. Public materials highlight multi-thousand-sample weekly capacity per instrumentation line and minimal input volumes.
- Data consistency: Olink's NPX framework supports plate-level normalization and cross-batch alignment with biological bridge samples. Practical success hinges on experimental layout and statistical post-processing rather than "raw speed."
- Why PEA fits cohorts: The assay's dual-antibody proximity design and DNA barcode readout deliver sensitivity for low-volume clinical samples, which is essential when CSF, plasma, or archived biobank aliquots are limited.
For a deeper primer on PEA-driven assay services and research-ready outputs, review Olink proteomics assay services explained.
Common challenges in large-scale cohort proteomics projects
Scaling proteomics is primarily a project execution challenge.
- Batch effects and long timelines: When cohorts run across months, drift accumulates. Without bridge samples on every batch and clear acceptance rules, cross-batch comparability deteriorates.
- Multi-center sample heterogeneity: Biobank and clinical sites differ in collection tubes, processing times, and storage histories; pre-analytical variation can overshadow biological signal if not harmonized.
- Limited volumes with high sensitivity needs: Cytokines and low-abundance immune markers may sit near LOD—pilot detectability checks are essential, especially for healthy cohorts.
- Milestone-driven schedules: Clinical programs often bind delivery to interim analyses. You need plate maps, randomization plans, and re-run rules prepared before the first plate is loaded.
Practical QC and batch-control framework for large-scale Olink proteomics
A robust framework combines experimental design with statistical normalization. Below is a compact, auditable recipe that maps to common Olink workflows.
Experimental layout
- Include biological bridge samples on every plate/batch (e.g., pooled cohort plasma). Aim for 8–16 bridges per plate for same-product alignment; expand the total count when bridging across products.
- Place Olink controls as specified by the platform, and randomize sample placement to avoid site or phenotype clustering on any single plate.
- Use consistent aliquoting volumes and record freeze–thaw counts; avoid >2 cycles when possible.
Normalization and cross-batch alignment
- Apply NPX plate/intensity normalization per Olink guidance. Compute assay-specific adjustments using the bridge samples relative to a reference plate.
- Validate success via CV reductions in controls, convergence of cluster structure in PCA/UMAP, and improved distribution overlap for adjusted assays.
Handling LOD and below-detect values
- Integrate LOD through OlinkAnalyze logic; retain NPX values with appropriate flags rather than blanket zeroing.
- Define panel-specific rules: exclude proteins undetected across all samples; consider imputation strategies only after confirming robustness in pilots.
Typical QC metric ranges (for planning)
| Metric | Planning range (cohort context) |
| Within-batch median CV (per analyte) | ~4–12% |
| Cross-batch median CV after bridging | ~8–20% |
| Bridge samples per plate (same product) | 8–16 |
| Detectability (healthy plasma, broad panel) | Often >75% overall; certain cytokines ≤50% |
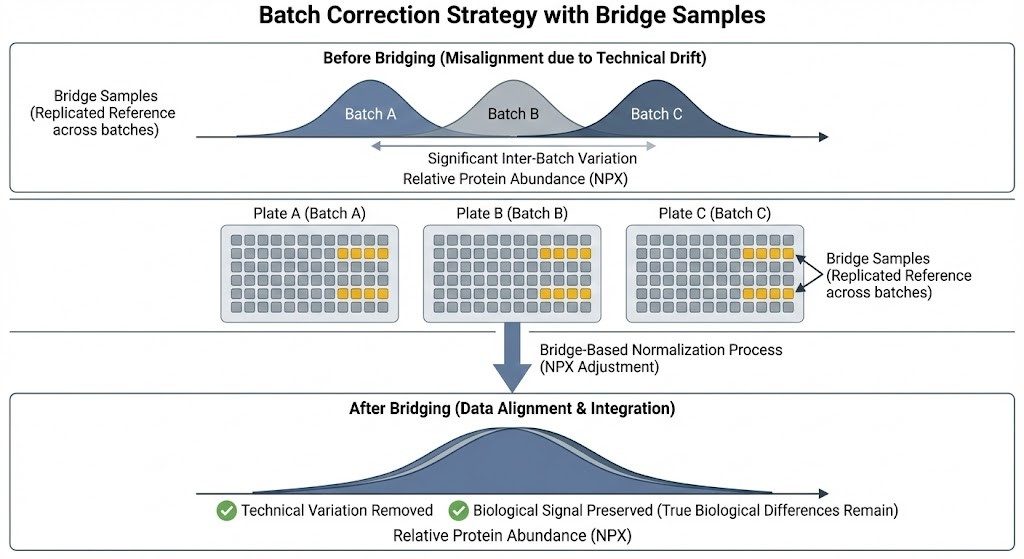 Batch correction strategy using biological bridge samples.
Batch correction strategy using biological bridge samples.
To mitigate technical variation in large-scale cohorts, overlapping biological bridge samples (gold) are included in every plate.
These ranges reflect commonly reported values across validation materials and cohort studies; confirm with a pilot on your matrices and panels. Olink's normalization and bridging practices are illustrated in the OlinkAnalyze vignettes and official data science resources.
For method context and service practices, see our internal explainer on Olink data analysis processes.
Operational robustness for multi-center and biobank samples
Operational consistency is as critical as analytical precision.
- Pre-analytical harmonization: Standardize collection SOPs (tube type, processing timeline), aliquoting, and storage at −80°C. Limit freeze–thaw cycles and document deviations.
- Shipping and chain of custody: Use dry-ice logistics, temperature monitors, and a sample manifest aligned to data governance requirements.
- Historical/biobank samples: Expect variability. Pilot runs should quantify detectability and CV before committing full batches; adjust panel selection for matrices like CSF or tissue lysates.
Practical handling recommendations are summarized in Olink proteomics sample submission guidelines.
Typical use scenarios for high-throughput cohorts
- Large immune-related cohorts (oncology, inflammation, autoimmunity): Useful for cytokine and chemokine landscapes; plan for variable detectability across sites.
- Clinical trial exploratory and companion studies: Discovery arms feeding into targeted validation; interim analyses require pre-baked QC acceptance criteria.
- Biobank population studies: Thousands of archived samples across years; bridge-heavy designs and robust normalization are essential.
- Two-stage discovery→validation programs: Start broad (Explore 3072/HT), down-select to focused targets, then validate with orthogonal assays.
For cohort design, power, and QC strategy at scale, see designing a large-scale study with Olink.
High-throughput clinical cohort immune profiling: why it's common and hard
Clinical cohort immune profiling is common—and challenging. Healthy or cytokine-poor cohorts often push detectability limits for Th2 cytokines (e.g., IL-4/IL-13). That doesn't preclude insight; it demands upfront pilots, careful LOD handling, and panel choices tuned to the population.
- Why it's prevalent: Immune signals underpin oncology, infection, autoimmunity, and neurology; proteomic profiles complement transcriptomics and clinical endpoints.
- From exploratory features to verified signals: Use discovery panels to identify candidate markers, then confirm with targeted immunoassays or MS.
If you're expanding into immune applications, review Olink immune profiling in translational research for practical pathways.
Integration with downstream validation and multi-omics pipelines
Treat proteomics as one layer in a convergent evidence chain.
- pQTL/proteogenomics: Link protein abundance to genetic variants and transcript levels to strengthen biological plausibility and clinical utility.
- Orthogonal validation: Confirm prioritized proteins using targeted MS or immunoassays; move to Focus/custom panels for locked-down verification.
- Study sequencing: Discovery (Explore HT/3072) → candidate triage (statistics + biology) → orthogonal validation → clinical utility assessment.
Population-scale precedents, such as the UK Biobank's Olink-based efforts, demonstrate feasibility and data value; see Olink's announcement on the expansion to Explore HT in UK Biobank's proteomics initiative.
Before you launch: a practical scoping checklist
Use this short checklist to align teams before committing to full-scale execution.
- Sample volume and matrices: Confirm minimal volume per assay (often 1–6 µL) and total reserve per sample; list allowable matrices.
- Project phase: Define discovery vs validation; estimate pilot size (e.g., 48–192 samples) to establish CV/detectability.
- Timelines and delivery nodes: Map interim analyses; specify batch counts and bridge sample numbers.
- Data integration and analysis: Plan NPX normalization, bridging, LOD handling, and multi-omics integration routes.
- Governance and auditability: Document SOPs, manifests, and QC thresholds; align with GCP/CAP/ISO governance expectations.
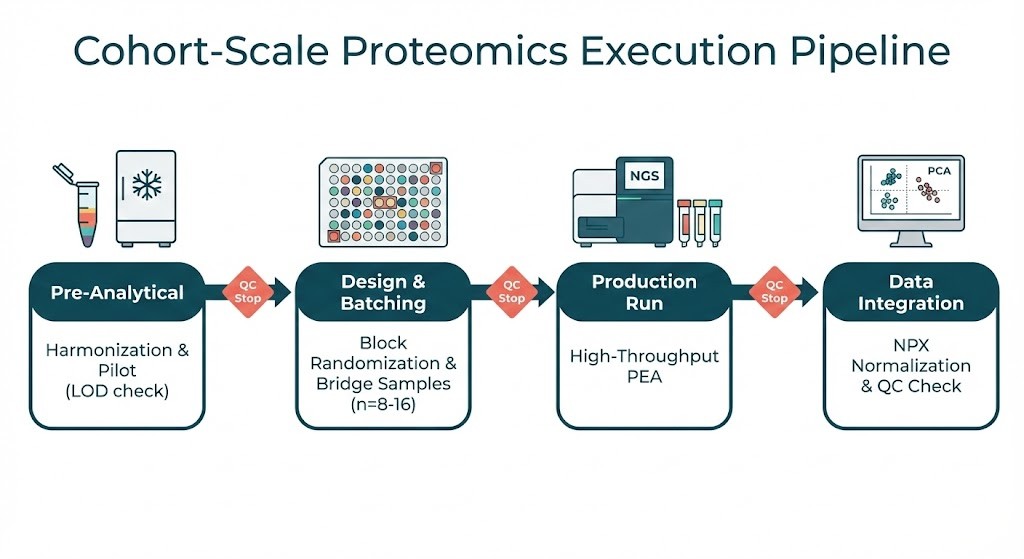 Execution workflow for large-scale clinical proteomics.
Execution workflow for large-scale clinical proteomics.
FAQ: High-throughput Olink proteomics for large cohort studies
How many samples can be processed in a high-throughput Olink project?
Cohort-scale designs frequently range from hundreds to >1,000 samples. Explore HT public materials indicate multi-thousand samples per week per instrumentation line when appropriately configured; align expectations via a pilot and capacity planning.
Is Olink proteomics suitable for multi-center cohort studies?
Yes—provided you harmonize pre-analytics, implement bridge samples on every batch, and apply NPX-based normalization with clear QC acceptance rules.
How are batch effects controlled in large-scale proteomics?
Through bridge samples, plate controls, and NPX normalization. Success is assessed by CV reduction, PCA/UMAP convergence, and distribution overlap across batches.
Can high-throughput Olink data support clinical cohort immune profiling?
Yes, but anticipate low detectability for certain cytokines in healthy/low-inflammation matrices. Pilot first, tune panels, and handle LOD rigorously.
How does high-throughput proteomics integrate with downstream validation?
Use proteomics for discovery, then confirm candidates via targeted MS or immunoassays, and integrate with pQTL/transcriptomics to build a robust evidence chain.
A neutral example mention — execution workflow
In a typical large-scale Olink engagement, an execution workflow might include plate randomization across sites, 8–16 biological bridge samples per plate, NPX normalization with bridge-based adjustments, diagnostics (CV, PCA), and a pilot to determine detectability in the target matrix. The goal is operational reproducibility, not platform promotion; these steps mirror standard practices documented in Olink materials and peer-reviewed studies.
Conclusion: enabling reliable proteomics at cohort scale
Large-scale Olink proteomics succeeds when you design for scalability and QC from day one. Treat bridge samples, normalization, and pre-analytics as core infrastructure; pilot for precision and detectability; and plan early for multi-omics integration and downstream validation. If you're preparing a cohort-scale project and want a grounded feasibility review, book a scoping call to align on design, QC thresholds, and timelines.
References and further reading
- Olink Explore HT overview and product documentation: Explore HT product page
- OlinkAnalyze software package (normalization and bridging vignettes): OlinkAnalyze on CRAN
- UK Biobank proteomics initiative (Explore HT expansion announcement): Olink news on UK Biobank
- PEA technology and service overview (Creative Proteomics): Olink proteomics assay services explained
- Large-scale design guidance (cohorts, power, QC strategy): Designing large-scale Olink proteomics studies




