Introduction: Revolutionizing Cardiovascular and Metabolic Disease Research
In the rapidly evolving field of medical research, Olink Proteomics has emerged as a transformative technology capable of uncovering complex molecular relationships in cardiometabolic diseases. By combining unprecedented sensitivity and high-specificity protein detection, this innovative approach enables scientists to decipher the intricate connections between lipid metabolism, insulin signaling, and cardiovascular inflammation at a previously unattainable resolution. The technology's capacity to analyze thousands of proteins from minute blood samples positions it as an indispensable tool for biomarker discovery and mechanistic understanding of diseases that constitute leading causes of global morbidity and mortality.
Cardiometabolic diseases, including type 2 diabetes, atherosclerosis, and hypertension, represent interconnected disorders with complex pathophysiology involving multiple biological systems. Traditional single-marker approaches have provided limited insights into the sophisticated network of molecular events underlying these conditions. Olink's proprietary Proximity Extension Assay (PEA) technology addresses this challenge by enabling simultaneous measurement of hundreds to thousands of proteins with exceptional accuracy, even at ultralow concentrations, thereby opening new avenues for understanding disease mechanisms and developing targeted interventions.
The Technological Advantage: How Olink PEA Transforms Protein Biomarker Research
- Unmatched Sensitivity and Specificity: Olink's PEA technology employs paired antibody probes labeled with complementary DNA strands that extend only when both antibodies bind to their target protein simultaneously. This dual-recognition mechanism minimizes false positives and enables detection of proteins at concentrations as low as fg/mL, which is crucial for measuring low-abundance inflammatory cytokines and metabolic regulators that play key roles in cardiometabolic diseases.
- Minimal Sample Requirements: The technology requires only 1-6 μL of biological samples (including plasma, serum, and tissue extracts), making it particularly valuable for longitudinal studies and research involving precious clinical cohorts where sample volume is often limited. This feature has enabled large-scale proteomic profiling across major biobanks, including the UK Biobank Pharma Proteomics Project, which aims to analyse proteins from 54,406 participants.
- Comprehensive Pathway Coverage: Olink offers specialized panels targeting cardiovascular, metabolic, inflammatory, and neurological diseases, each containing carefully selected proteins that represent key biological pathways. These targeted approaches provide deep insights into specific physiological systems while enabling the discovery to novel biomarker relationships through cross-panel analyses.
Application Case Studies in Cardiometabolic Disease Research
1. Uncovering Inflammatory Pathways in Cardiovascular Disease
- A study published on Circulation (Wijk et al., 2020) investigates the role of systemic inflammation in mediating the relationship between comorbidity burden (e.g., hypertension, diabetes) and cardiac dysfunction in HFpEF. Using Olink proteomics, the study quantified 248 circulating proteins via multiplex immunoassays, enabling a comprehensive analysis of inflammatory pathways. Using both deductive (principal component analysis of a priori inflammation proteins) and inductive (weighted coexpression network analysis) approaches, the researchers identified key inflammation-related protein clusters, including those centred on TNFR1 and IGFBP7, which were associated with worsened hemodynamics and right ventricular function.
- Researchers (Zhao et al., 2024) employ Olink technology to profile 92 immune response-related proteins in serum samples, identifying 59 differentially expressed proteins between ischemic stroke patients and controls.By leveraging Olink's high-sensitivity multiplex assays, the study pinpointed six key biomarkers (e.g., MASP1, CLEC4D) involved in inflammatory pathways, including C-type lectin receptor signaling and the PI3K-AKT pathway, which are critical in stroke pathogenesis. The Olink-based diagnostic model demonstrated high accuracy (AUC up to 0.954) in distinguishing IS cases, highlighting how Olink proteomics enables the discovery of inflammation-driven mechanisms in cerebrovascular disease and contributes to the broader theme of inflammatory pathways in cardiovascular disorders.
2. Deciphering Insulin Signaling Defects in Type 2 Diabetes
A study leverages a soft-clustering (archetype) analysis to characterise the etiological heterogeneity in newly diagnosed type 2 diabetes (T2D), identifying four distinct archetypes that reflect varying degrees of insulin signalling defects, including insulin resistance (prominent in archetypes C and D) and β-cell dysfunction (evident in archetype A). By integrating multi-omics data, the research utilized Olink proteomics panels for high-throughput plasma protein profiling, which was instrumental in deciphering circulating biomarkers—like inflammatory cytokines (e.g., IL-8, IL-18) and metabolic regulators (e.g., IGFBPs)—that are strongly associated with these archetypes and their underlying genetic risk scores.
 Figure 1: Clinical characteristics of the four archetypes, and groups with archetype scores identified at the extremes of the baseline phenotype spectrum. (Wesolowska-Andersen, A. et al., 2022)
Figure 1: Clinical characteristics of the four archetypes, and groups with archetype scores identified at the extremes of the baseline phenotype spectrum. (Wesolowska-Andersen, A. et al., 2022)
3. Integration with Metabolic Profiling: Multi-Omic Insights
Researchers leverage Olink's innovative PEA-NGS (Proximity Extension Assay with Next Generation Sequencing) platform to enable comprehensive integration of plasma proteome profiling with metabolic and clinical data, offering multi-omic insights into health and disease dynamics. By analyzing nearly 1,500 proteins in longitudinal cohorts, including healthy individuals and newly diagnosed type 2 diabetes (T2D) patients, the research demonstrates robust correlations between protein levels and key metabolic parameters such as BMI, fasting glucose, and lipid profiles. Olink's high-sensitivity, multiplexed approach facilitates the identification of protein signatures associated with metabolic dysregulation, such as differential expression in non-obese T2D subgroups and stratification of patients based on metformin treatment response, underscoring its pivotal role in deriving actionable multi-omic insights for precision medicine.
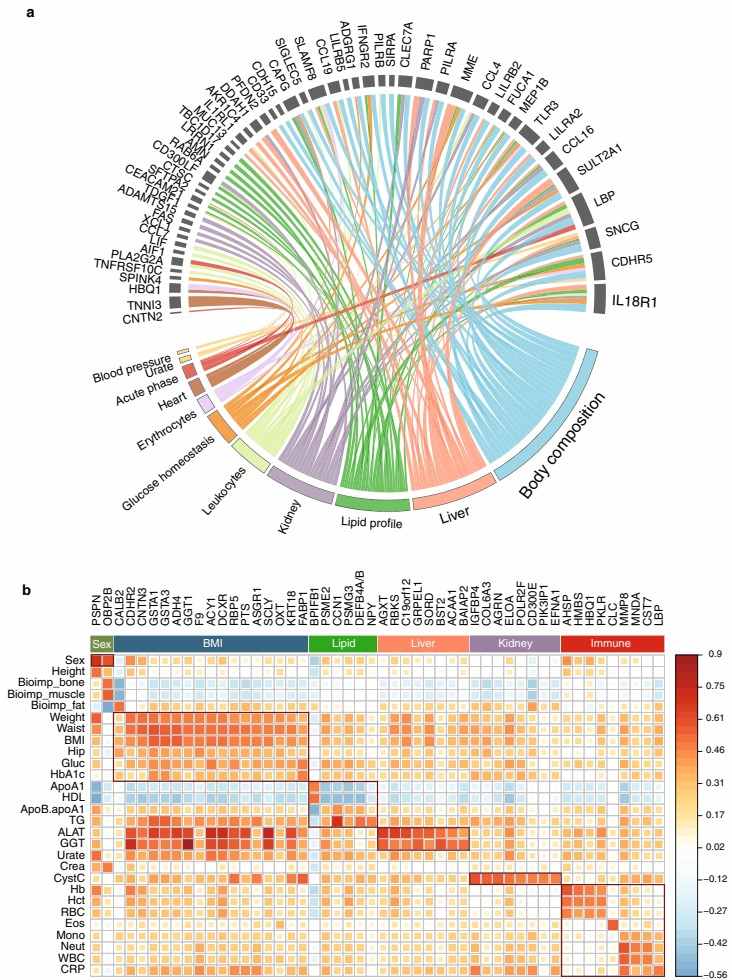 Figure 2: Associations of proteins with clinical assays. (Zhong, W. et al., 2021)
Figure 2: Associations of proteins with clinical assays. (Zhong, W. et al., 2021)
4. From Association to Mechanism: Experimental Validation of Proteomic Discoveries
A study (Woolley et al., 2021) employs Olink's high-throughput proteomics technology to profile 363 proteins in 429 patients with Heart Failure with Preserved Ejection Fraction (HFpEF). The initial association was established through unsupervised machine learning, leveraging high-quality protein data generated by Olink's PEA to identify four distinct patient clusters with unique biomarker profiles and clinical outcomes. Critically, the research then progressed to a mechanistic exploration: differential expression analysis of the Olink-derived protein signatures revealed cluster-specific dysregulated pathways, such as inflammatory activation in the diabetic/renal cluster and PI3K/AKT signaling in the cluster with ischemic/COPD comorbidity.
Thus, Olink technology was instrumental not only in identifying protein-based subgroups but also in providing the precise proteomic data necessary to hypothesize about the underlying pathophysiological mechanisms driving HFpEF heterogeneity.
Future Directions: Toward Personalized Cardiometabolic Medicine
As Olink technologies continue to evolve, their applications in cardiometabolic medicine are expanding in several promising directions:
- High-Throughput Population Screening: The ongoing UK Biobank Pharma Proteomics Project exemplifies how large-scale protein profiling can identify novel biomarkers for disease risk stratification and prevention. This resource will enable researchers to examine prospective relationships between thousands of plasma proteins and incident cardiometabolic diseases in a cohort of over 50,000 participants.
- Drug Development Applications: Olink proteomics is increasingly being deployed throughout the drug discovery pipeline, from target identification and validation to patient stratification and pharmacodynamic biomarker development. The ability to monitor responses to therapeutic interventions at the pathway level provides unprecedented insights into drug mechanisms of action and individual variability in treatment responses.
- Single-Cell and Tissue Proteomics: Emerging applications of Olink technology at the single-cell level and in tissue specimens promise to resolve cellular heterogeneity within metabolic organs such as pancreas, liver, and adipose tissue. These advances may reveal how protein expression and signaling differ between cell types in the context of cardiometabolic disease.
Conclusion: Transforming Our Approach to Cardiometabolic Diseases
Olink Proteomics has fundamentally transformed our ability to investigate the complex molecular landscape of cardiometabolic diseases. By providing highly sensitive, specific, and multiplexed protein measurements, this technology has enabled researchers to move beyond individual biomarkers to understand interconnected biological pathways. The insights from Olink studies have revealed novel aspects of dysregulated lipid metabolism, insulin signaling, and inflammatory pathways in conditions ranging from type 2 diabetes to ischemic heart disease.
As the technology continues to evolve and integrate with other omics approaches, it promises to accelerate the development of personalized prevention strategies and targeted therapies for cardiometabolic disorders. The progression from biomarker discovery to mechanistic understanding represents a paradigm shift in how we approach these complex diseases, offering hope for more effective and individualized interventions in the future.
FAQs
1. How does Olink technology help in understanding the mechanisms of cardiometabolic diseases?
Olink proteomics moves beyond simple biomarker identification to elucidate disease mechanisms by enabling:
- Multi-omics Integration: By combining protein data with genomic and metabolomic data, researchers can build a more complete picture of disease biology.
- Pathway Analysis: The proteins in Olink panels are linked to specific biological pathways. When a study finds a set of proteins from such a panel to be dysregulated, it directly suggests the activation or disruption of these critical pathways in the disease state.
- Identification of Causal Relationships: The high-quality protein data from Olink, when combined with genetic information (e.g., through pQTL studies or Mendelian randomization), can help determine if an associated protein is likely to be a cause or a consequence of the disease, which is vital for identifying potential drug targets.
2. What are the practical considerations for designing a study using Olink proteomics for cardiometabolic research?
Key considerations when planning a study include:
- Panel Selection: Olink offers various targeted panels. For cardiometabolic research, the obvious choice is the Cardiometabolic panel. However, other panels like Inflammation, Cardiovascular II/III, or Organ Damage are also highly relevant, as inflammation and organ damage are core components of these diseases. The choice can be based on:
- Disease Name Matching: Directly selecting a panel based on the disease of interest.
- Pathway Correspondence: Selecting a panel that covers biological pathways of interest (e.g., immune response).
- Literature Review: Searching existing literature to identify specific proteins of interest and then choosing a panel that contains them.
- Study Design and Cohort Size: The technology is well-suited for large cohort studies, as demonstrated by its use in the UK Biobank and the Chinese CKB cohort. Its low sample volume requirement is a significant advantage for such studies.
- Platform Comparison: Researchers should be aware that different proteomic platforms (e.g., Olink vs. SomaScan) may show moderate correlations for the same protein. A large-scale comparison study in the CKB cohort found that while correlations were often moderate, the platforms showed complementary strengths in identifying disease-associated proteins, suggesting that the choice of platform can influence results.
Related Reading
For deeper insights, we recommend exploring the following related guides:
Olink Proteomics in Clinical Research: A Game Changer — A demonstration of how high-plex proteomics advances precision medicine, validates novel biomarkers, and elucidates disease mechanisms through sensitive protein profiling.
The Future of Proteomics: Where Olink Fits Into Emerging Technologies — A perspective on how Olink's platform, by merging PEA with NGS, streamlines biomarker and target validation, defining precision medicine pathways through scalable proteomics.
References
- Sanders-van Wijk, S., Tromp, J., Beussink-Nelson, L., Hage, C., Svedlund, S., Saraste, A., et al. (2020). Proteomic Evaluation of the Comorbidity-Inflammation Paradigm in Heart Failure With Preserved Ejection Fraction. Circulation, 142(21), 2029-2044.
- Zhao, T., Zeng, J., Zhang, R., Fan, W., Guan, Q., Wang, H., et al. (2024). Serum Olink Proteomics-Based Identification of Protein Biomarkers Associated with the Immune Response in Ischemic Stroke. Journal of Proteome Research, 23(3), 1118-1128.
- Wesolowska-Andersen, A., Brorsson, C. A., Bizzotto, R., Mari, A., Tura, A., Koivula, R., et al. (2022). Four groups of type 2 diabetes contribute to the etiological and clinical heterogeneity in newly diagnosed individuals: An IMI DIRECT study. Cell Reports Medicine, 3(1), 100477.
- Zhong, W., Edfors, F., Gummesson, A. et al. Next generation plasma proteome profiling to monitor health and disease. Nat Commun 12, 2493 (2021).
- Woolley, R. J., Ceelen, D., Ouwerkerk, W., Tromp, J., Figarska, S. M., Anker, S. D., et al. (2021). Machine learning based on biomarker profiles identifies distinct subgroups of heart failure with preserved ejection fraction. European Journal of Heart Failure, 23(6), 983-991.




