Introduction: The Need for Advanced Proteomics in Immunology Research
The immune system represents one of the most complex biological networks in human biology, characterized by dynamic protein interactions that dictate inflammatory responses, immune cell communication, and disease pathogenesis. Traditional protein detection methods have faced significant limitations in comprehensively capturing this complexity due to constraints in multiplexing capacity, sensitivity, and sample volume requirements. The introduction of high-plex proteomic technologies like Olink Explore 1536 has revolutionized how immunology researchers and pharmaceutical scientists approach the study of inflammatory diseases, immune responses, and therapeutic development.
- Olink Explore 1536 employs Proximity Extension Assay (PEA) technology, which enables simultaneous measurement of up to 1,536 proteins from minimal sample volumes (as little as 1-6μL of plasma or other biofluids) . This technological advancement is particularly valuable for immunology research, where protein biomarkers often exist at low concentrations but carry critical information about disease states and therapeutic responses. The platform's extensive coverage of immune-related pathways and inflammatory mediators provides researchers with an unprecedented ability to map protein networks underlying immunological processes.
This article will explore how Olink Explore 1534 is advancing immunology and inflammation research through key applications, including mapping immune pathway networks, identifying critical protein signatures in inflammatory diseases and integration with multi-omics approaches.
Comprehensive Coverage of Immune Pathways and Inflammatory Mediators
The Olink Explore 1536 platform provides exceptional coverage of proteins across critical immune and inflammatory pathways, enabling researchers to obtain a systems-level view of immune responses.
1. Broad Pathway Coverage: The platform covers 100% of major signaling pathways involved in immune responses, including cytokines, chemokines, growth factors, and their receptors. This extensive coverage allows researchers to monitor complete biological cascades rather than isolated components. The technology detects proteins across a dynamic range of 10 logs, which is essential for measuring both high-abundance acute phase proteins and low-abundance signaling cytokines that are functionally important in immune responses.
 Figure 1: Principle of the PEA-NGS method and description of cohorts. (Zhong, W. et al., 2021)
Figure 1: Principle of the PEA-NGS method and description of cohorts. (Zhong, W. et al., 2021)
2. Key Immune Protein Classes: The Olink Explore 1536 platform includes panels designed to measure a broad range of proteins implicated in immunological processes. These panels provide comprehensive coverage of key immune-related protein families, such as interleukins, interferons, tumor necrosis factors, chemokines, and immune checkpoint regulators. This approach enables researchers to simultaneously monitor a wide spectrum of relevant biomarkers involved in immune activation and regulation.
3. Inflammation-Focused Panels: The Olink Explore 1536 panel facilitates focused analysis of inflammatory processes in long COVID, measuring proteins linked to type II interferon signaling (e.g., IFN-γ, CXCL9), NF-κB pathway components (e.g., TNF, IL-6), and neutrophil activation markers (e.g., ANXA3). This specialized profiling is particularly valuable for identifying inflammatory PASC subcategories, where these pathways drive persistent immune dysregulation.
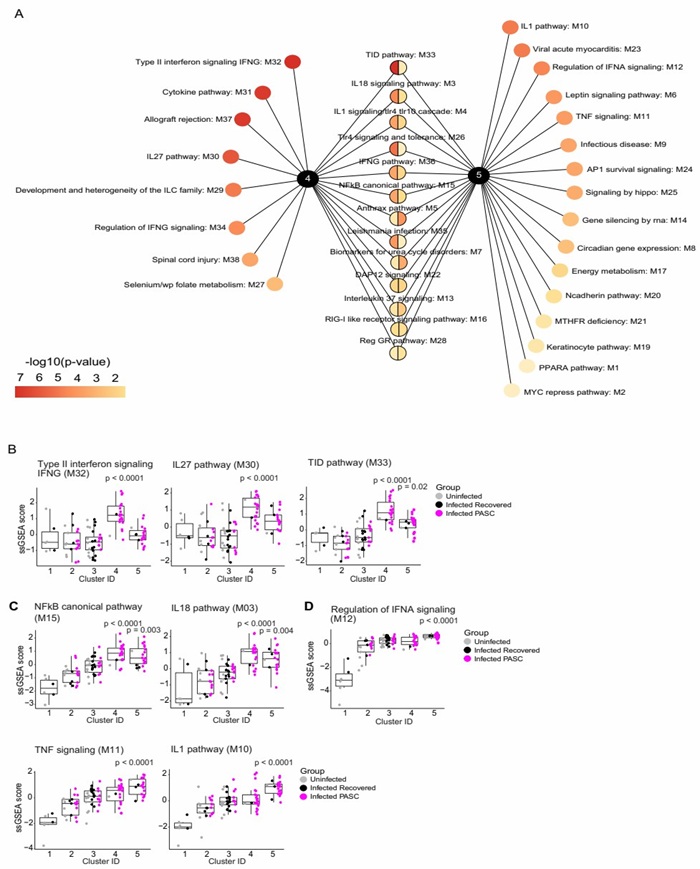 Figure 2: Key pathway modules driving inflammatory PASC signatures. (Talla, A. et al., 2023)
Figure 2: Key pathway modules driving inflammatory PASC signatures. (Talla, A. et al., 2023)
The platform's design incorporates deep biological insight into immune system function, ensuring that the measured proteins provide meaningful information about immune status, activation, and regulation.
Mapping Cellular Signaling in Immune Responses and Inflammation
Olink Explore 1536 enables detailed mapping of immune cell signaling and inflammatory responses through its ability to simultaneously quantify multiple components of signaling pathways.
- Cytokine Signaling Networks: Research using the Olink Explore 1536 platform has elucidated complex cytokine signaling networks that characterize an inflammatory subset of long COVID patients. For example, the analysis identified type II interferon (IFN-γ) signaling and canonical NF-κB pathways, particularly driven by TNF, as the most differentially enriched, defining clusters with persistent inflammation. Additionally, proteins including CCL7 and S100A12 were highlighted in a diagnostic panel, underscoring the utility of proteomic signatures in stratifying PASC. (Talla, A. et al., 2023)
- Transcriptional Regulation of Immune Genes: The integration of proteomic data from Olink Explore 1536 with transcriptional profiling from in vivo CRISPR screens has elucidated how interferon (IFN) signaling pathways, mediated by transcription factors like STAT1 and IRF3, coordinate immune evasion in cancer. This IFN-responsive network represents a critical nodal point in immunotherapy resistance that can be quantitatively monitored through its downstream target proteins, measurable at scale using the Olink Explore 1536 platform, enabling comprehensive profiling of inflammatory and immune-related biomarkers in patient sera. (Dubrot, J. et al., 2022)
- Cancer-Related Inflammation: In oncology, the platform helps decipher the interplay between cancer and the immune system. One study identified cancer-specific protein signatures and found that immune-cell-related cancers shared a large number of upregulated proteins, many of which were associated with immune-related functions.
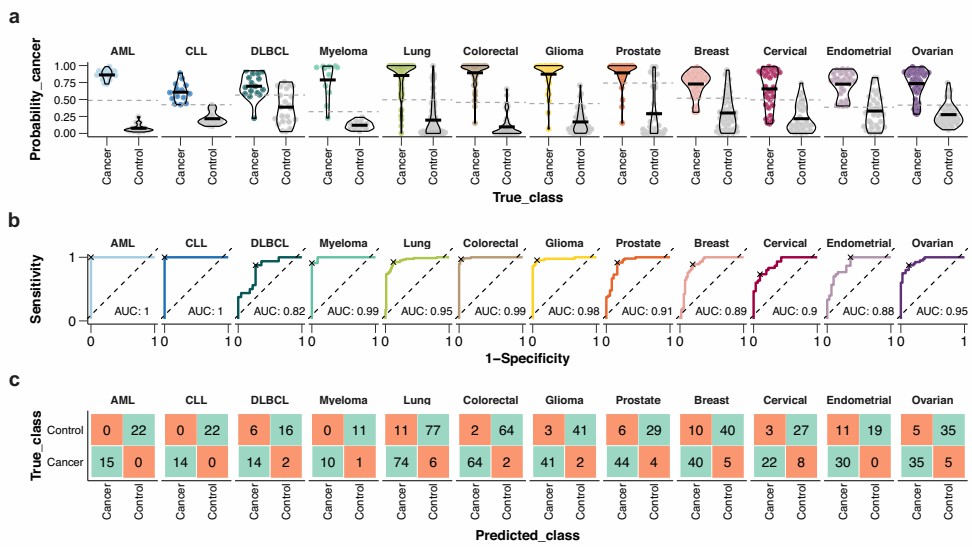 Figure 3: Performance of the classification models for each cancer on the test set. (Álvez, M.B. et al., 2023)
Figure 3: Performance of the classification models for each cancer on the test set. (Álvez, M.B. et al., 2023)
These insights demonstrate how high-plex protein data can reveal the architecture of immune signaling networks and identify key regulatory nodes that might be targeted for therapeutic intervention.
Application Case Studies in Immunology and Inflammation Research
Olink Explore 1536 facilitates the translation of proteomic findings into clinically relevant insights, from identifying biomarkers to understanding disease mechanisms.
Biomarker Discovery for Disease Stratification
Using the Olink Explore 1536 platform, proteomic analysis of 1,459 plasma proteins in 47,151 UK Biobank participants identified biomarkers such as THBS2 and IGFBP7 that significantly differentiate NAFL from cirrhosis. These proteins enabled robust disease stratification, outperforming models based on liver enzymes and genetic risk scores in discrimination accuracy.
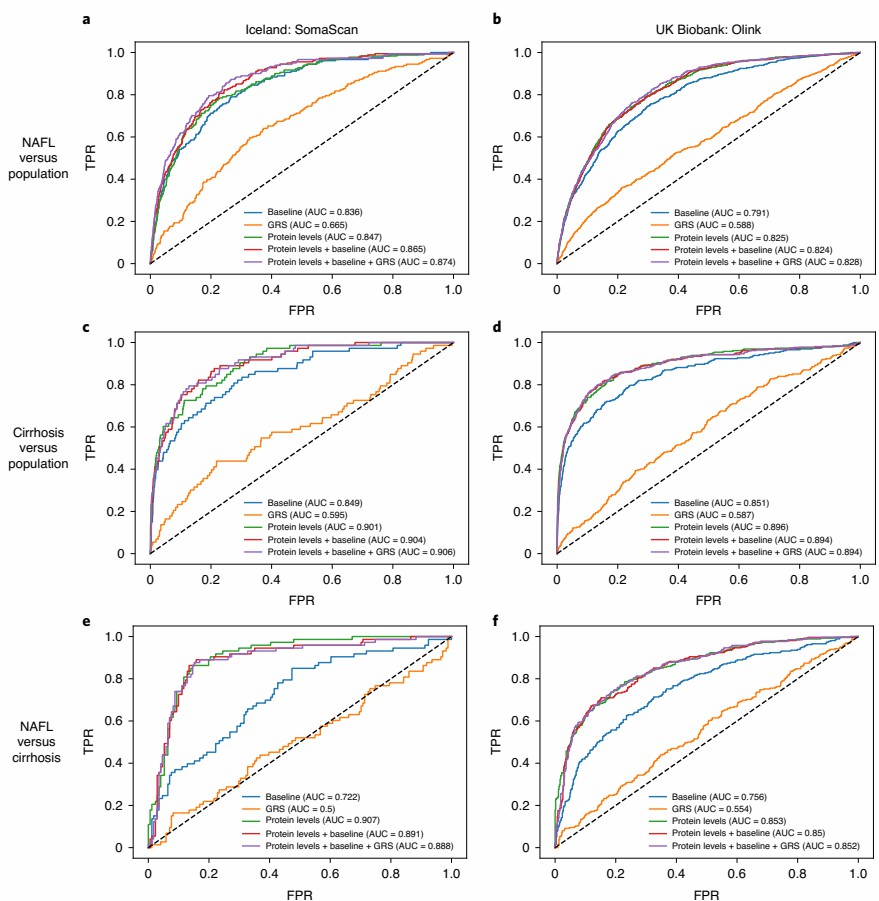 Figure 4: ROC for models trained to discriminate between NAFL and cirrhosis. (Sveinbjornsson, G. et al., 2022)
Figure 4: ROC for models trained to discriminate between NAFL and cirrhosis. (Sveinbjornsson, G. et al., 2022)
Mechanistic Insights into Disease Pathobiology
The integration of Olink Explore 1536 proteomic data from 1,180 individuals revealed 256 novel pQTLs, providing mechanistic insights into disease pathobiology. For instance, a cis-pQTL for gastrin-releasing peptide (GRP) demonstrated a shared genetic signal with type 2 diabetes risk, suggesting a role in appetite regulation and body weight. This highlights how proteogenomics can identify new protein-disease links.
The protein-disease network generated from the colocalization analysis, further visualizes these connections, linking 224 protein targets to 575 unique health outcomes and illustrating the platform's power to map biological pathways. (Koprulu, M. et al., 2023)
Therapeutic Monitoring
The platform is suitable for monitoring protein levels in clinical studies. A study leveraged the Olink Explore 1536 platform to measure ~3,000 plasma proteins in 41,931 UK Biobank participants. Sparse signatures of 5-20 proteins significantly improved 10-year risk prediction for 67 diseases, surpassing models based on clinical data alone. The findings demonstrate the utility of proteomic signatures for enhancing therapeutic monitoring and early disease detection.
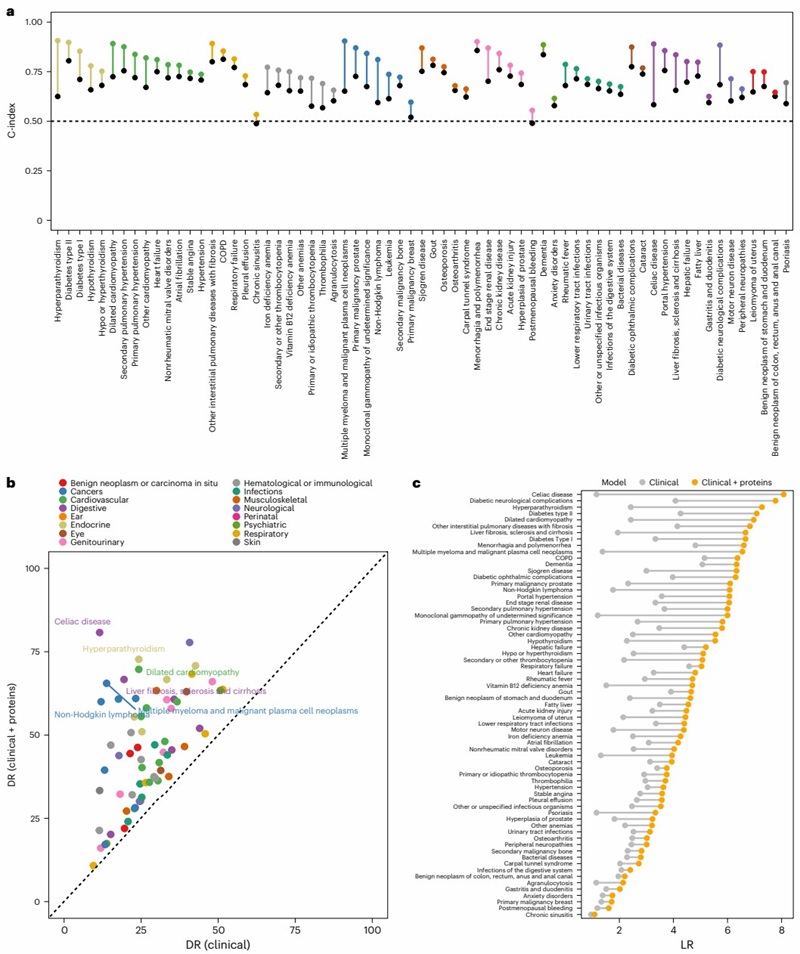 Figure 4: Improvement in predictive performance of disease incidence by addition of proteomic information on top of basic clinical risk factors for 67 diseases. (Carrasco-Zanini, J. et al., 2024)
Figure 4: Improvement in predictive performance of disease incidence by addition of proteomic information on top of basic clinical risk factors for 67 diseases. (Carrasco-Zanini, J. et al., 2024)
Integration with Other Data Types
Olink Explore 1536 is increasingly used as part of integrated multi-omics strategies that provide a more comprehensive understanding of immune system function.
- Proteogenomic Integration: The combination of genomic and proteomic data enables identification of protein quantitative trait loci (pQTLs), which reveal how genetic variation influences protein abundance. This approach is particularly powerful in immunology, where many disease-associated genetic variants affect immune function through protein regulation.
- Proteo-transcriptomic Correlations: Simultaneous analysis of transcriptomic and proteomic data reveals important insights about immune regulation. That mRNA abundance does not accurately predict protein levels underscores the importance of direct protein measurement for understanding immune function.
- Temporal Dynamics of Immune Responses: The integration of Olink data with other omics layers across time series enables researchers to build dynamic models of immune responses. Such temporal data is crucial for understanding the progression of immune responses and inflammatory processes.
These integrated approaches demonstrate how Olink proteomics contributes to systems immunology, helping researchers build comprehensive models of immune function in health and disease.
Conclusion: The Future of Immunoproteomics with Olink Explore 1536
Olink Explore 1536 has established itself as a transformative technology in immunology and inflammation research, enabling unprecedented insights into protein networks underlying immune responses. The platform's sensitivity, specificity, and comprehensive coverage of immune pathways make it particularly valuable for unraveling the complexity of inflammatory diseases, immune cell communication, and host-pathogen interactions.
- As the field advances, several developments promise to further enhance the utility of Olink Explore in immunology research. The growing public protein databases provide rich context for interpreting new findings.
- The application of machine learning algorithms to high-plex protein data is yielding sophisticated models for disease classification, prognosis prediction, and treatment selection.
- Furthermore, the technology's compatibility with minimal sample volumes positions it ideally for precious longitudinal samples in clinical trials and pediatric immunology studies.
The future of immunology research lies in embracing these high-plex, high-precision technologies that can capture the complexity of immune responses while providing practical advantages for translational applications. Olink Explore 1536 represents a significant step toward this future, empowering researchers to decode the intricate language of immune proteins and harness this knowledge for therapeutic advancement.
FAQs
1. What is Olink Explore 1536 and why is it valuable for immunology research?
Olink Explore 1536 is a high-throughput proteomics platform that allows for the simultaneous measurement of 1,536 proteins from a very small sample volume (as low as 1-4 µl of plasma or other body fluids). Its value in immunology research stems from several key advantages.
- High-Plex Biomarker Panels: The platform includes pre-configured panels targeting proteins across major biological pathways, with dedicated panels for immunology, inflammation, and neurology, providing comprehensive coverage of immune-relevant proteins.
- Exceptional Sensitivity and Dynamic Range: It can detect very low-abundance proteins (in the fg/mL range), which is crucial for measuring key inflammatory cytokines and signaling molecules that are often present at minimal levels. Its wide dynamic range allows it to capture both high- and low-abundance proteins in a single run.
- Ideal for Liquid Biopsy: The technology is particularly suited for analyzing blood-based samples, enabling minimally invasive "liquid biopsies" to monitor immune responses and inflammation over time.
2. How does Olink Explore 1536 facilitate multi-omics integration in immunology?
A major strength of Olink Explore 1536 is its foundation on Next-Generation Sequencing (NGS) for readout. This creates a powerful synergy with other NGS-based technologies:
- Discovery of pQTLs: By combining genomic data with protein data from the same individuals, researchers can identify protein quantitative trait loci (pQTLs). pQTLs are crucial for understanding the genetic regulation of the immune response and for using Mendelian randomization to infer causal relationships between proteins and diseases.
- Linking Transcriptomics and Proteomics: The ability to use similar NGS workflows for both RNA sequencing (e.g., single-cell transcriptomics) and protein measurement makes it easier to perform integrated analyses.
Related Reading
For deeper insights, we recommend exploring the following related guides:
How Olink's Cytokine Panel Accelerates Inflammation Research — An overview of how targeted proteomics deepens mechanistic understanding of immune pathways and validates novel protein signatures across diverse inflammatory diseases through high-sensitivity multiplex profiling.
Exploring the Olink Inflammation Panel: Key Features & Applications — This article delves into how the panel's high-sensitivity PEA technology empowers immunology research by enabling large-scale validation of key inflammatory protein signatures, thereby refining disease stratification and accelerating the development of targeted therapeutic strategies.
References
- Zhong, W., Edfors, F., Gummesson, A. et al. Next generation plasma proteome profiling to monitor health and disease. Nat Commun 12, 2493 (2021).
- Talla, A., Vasaikar, S.V., Szeto, G.L. et al. Persistent serum protein signatures define an inflammatory subcategory of long COVID. Nat Commun 14, 3417 (2023).
- Dubrot, J., Du, P.P., Lane-Reticker, S.K. et al. In vivo CRISPR screens reveal the landscape of immune evasion pathways across cancer. Nat Immunol 23, 1495–1506 (2022).
- Álvez, M.B., Edfors, F., von Feilitzen, K. et al. Next generation pan-cancer blood proteome profiling using proximity extension assay. Nat Commun 14, 4308 (2023).
- Sveinbjornsson, G., Ulfarsson, M.O., Thorolfsdottir, R.B. et al. Multiomics study of nonalcoholic fatty liver disease. Nat Genet 54, 1652–1663 (2022).
- Koprulu, M., Carrasco-Zanini, J., Wheeler, E. et al. Proteogenomic links to human metabolic diseases. Nat Metab 5, 516–528 (2023).
- Carrasco-Zanini, J., Pietzner, M., Davitte, J. et al. Proteomic signatures improve risk prediction for common and rare diseases. Nat Med 30, 2489–2498 (2024).








