Inflammation plays a central role in many basic biology questions—from immune-cell signalling to tissue repair and ageing. For researchers, characterizing dozens of inflammation mediators simultaneously can uncover signalling networks, regulatory feedback loops, or responses to experimental perturbations. The Olink Inflammation Panel, built on Olink proteomics technology, enables such multiplex profiling with high sensitivity and small sample volumes. This article gives an in-depth view of its technical foundation, versions, strengths, limitations, and real-world applications in non-clinical biology settings.
What Is the Olink Inflammation Panel? Components & Biomarker Coverage
The Olink inflammation panel is a multiplex protein assay designed to measure many inflammation-related proteins (cytokines, chemokines, soluble receptors, and acute-phase proteins) in a single experiment.
Key attributes:
Protein set: Typically ~90-92 biomarkers, including both pro-inflammatory mediators (e.g., IL-6, TNF-α, IL-1β) and regulatory/anti-inflammatory ones (e.g., IL-10).
Sample type & volume: Works with plasma or serum. Requires very low volume—≈1 μL sample per panel in many configurations.
Quantitation metric: Data is reported as NPX (Normalized Protein eXpression), a log2-scaled, relative quantification value. It allows comparison across samples but does not give absolute concentrations unless paired with calibration or special variants.
Breadth of coverage: Alongside cytokines and chemokines, it includes growth factors, soluble receptors, and acute-phase reactants, enabling mapping of multiple arms of inflammation: signalling, recruitment, and resolution.
This biomarker coverage enables studies of immune activation, signalling cascades, response to stimuli, ageing, or environmental impacts, where multiple mediators interact.
Underlying Technology: How Olink's PEA Powers Sensitivity & Specificity
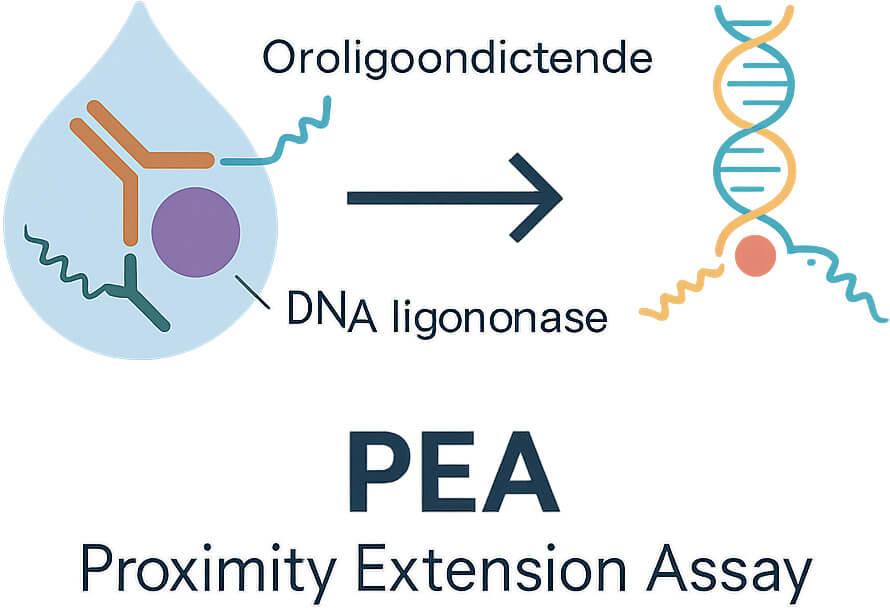
Understanding the Proximity Extension Assay (PEA) is central to appreciating why the Olink inflammation panel performs well in low sample volumes and for low-abundance proteins.
How PEA works:
Each target protein is bound by two specific antibodies, each conjugated to unique oligonucleotides. When both antibodies bind the same target, the oligos are brought into proximity.
These oligos hybridize and are extended by a DNA polymerase to form a DNA barcode. That barcode is then amplified (by qPCR or Next Generation Sequencing readout) and quantified.
Because both binding events are needed, background noise is reduced; cross-reactivity is less of a problem compared to single‐antibody methods.
Performance metrics:
- High specificity and low background, owing to dual recognition + DNA coupling.
- Wide dynamic range, allowing detection of proteins from relatively high to very low abundance.
- Good reproducibility across runs; low coefficient of variation for intra-assay precision.
A peer-reviewed study, Technical performance of a proximity extension assay inflammation panel, rigorously assessed the performance of Olink's inflammation panels. It showed robust linearity, high detectability across many proteins, and meaningful dynamic range even in complex matrices.
Another comparative evaluation of the Olink Target 48 Cytokine panel vs MSD & Luminex platforms demonstrated that Olink requires only ~1 μL plasma vs much larger volumes for bead-based or electrochemiluminescence platforms, while maintaining comparable or better precision for overlapping targets.
Versions & Panel Sizes: Which One Fits Your Research
Olink offers multiple panel "plex" sizes and flexible/customised panels. Choosing the right version depends on the number of analytes, sample volume, throughput, and research question.
| Panel Type | Approx. Number of Markers | Sample Volume | Throughput / Suitability |
| Target 48 / Target 96 | ~48 or ~92 markers | ~1 μL plasma/serum | Medium plex; useful when you want a broad yet manageable protein set. |
| Explore / High-plex (e.g., 384, 3072) | Hundreds to thousands of markers | Still low (but somewhat higher) | Use when you need wide discovery / system profiling. |
| Flex / Custom Panels | ~5-30 markers (user-chosen) | ~1 μL | Best when you already have strong hypotheses or a limited sample; selecting the most relevant inflammation‐related proteins. |
Olink Flex, for example, allows combining 15-21 human inflammation-related proteins into a custom panel from a library of ~200 validated targets. It offers results in both absolute (pg/mL) and relative (NPX) quantification.
Sample Types, Data Output & Interpretation Basics
Before running the panel, careful planning of sample collection, processing, and data interpretation is crucial for valid results.
Sample-related considerations:
Use plasma or serum; ensure consistent sample type across all your experiments (plasma vs serum differences can introduce artefacts).
Pre-analytic handling matters: time to freeze, freeze-thaw cycles, storage temperature. Inflammatory proteins are often sensitive to handling.
Volume: minimal (often ~1 µL), but ensure you have enough replicates and/or backup.
Data readouts and QC:
- NPX values are log2-transformed relative expression. Negative NPX values are possible (low abundance), but trends and differences are meaningful.
- Important QC metrics include limit of detection (LOD), lower & upper limit of quantification (LLOQ/UQO), intra- and inter-assay coefficient of variation (CV), and missingness for low abundance proteins.
- Normalisation: Olink has built-in internal controls; but data still often needs downstream normalisation (batch effects, plate effects) in experimental design and analysis.
Interpretation cautions:
- The NPX is relative expression—cannot compare directly with absolute concentration unless calibration has been performed.
- Low abundance proteins near LOD may show more variability or missing values.
- Multiple hypothesis testing: when many proteins are tested, control for false discovery (e.g., FDR correction) is essential.
For more on the data processing steps, you might refer to "Understanding Olink's Data Analysis Process: From Raw Data to Insights," which walks through NPX normalization, QC thresholds, and filtering.
Advantages & Limitations of the Olink Inflammation Panel
No method is perfect; being aware of the pros and cons helps in planning, interpreting, and reporting experiments.
Advantages:
High sensitivity even with tiny sample volumes; ideal for precious or limited samples.
Multiplexing many inflammation markers simultaneously saves time, cost, and sample.
Dual antibody + DNA barcode architecture reduces background and enhances specificity.
Reproducibility across plates and labs: well-validated assays enable comparing across batches if QC is consistent.
Flexibility especially with Flex panels: you can tailor marker sets to your hypothesis.
Limitations:
Relative quantification: NPX ≠ absolute concentration (unless specific calibration) which can complicate comparing across studies or platforms.
Dynamic range issues: very high abundance proteins may saturate, very low ones near detection limits may be noisy.
Cost: multiplex panels, especially high-plex and discovery-level, can have significant reagent and instrument costs.
Dependency on antibody performance: although many are well validated, some targets may still show cross-reactivity or suboptimal detection.
Applications in Non-Clinical Research
Here are examples of how the Olink inflammation panel has been used in academic / translational / R&D settings (non-clinical) to generate insights.
Technical Performance Study
The paper Technical performance of a proximity extension assay inflammation panel assessed the ability of Olink's inflammation panel to detect proteins in human plasma. They reported good linearity, high detectability for most proteins, and reliable dynamic range in complex matrices.
Platform Comparison: Olink vs MSD / Luminex
A comparative study of the Olink Target 48 Cytokine panel vs Mesoscale Discovery (MSD) and Luminex found that Olink required far smaller sample volume (~1 µL vs ~12–20 µL) while yielding similar precision and detectability for shared proteins. This is especially valuable for studies where samples are limited.
Custom Panel Use: Olink Flex
Olink Flex enables selecting a set of ~15-21 inflammation-related proteins for situations when you want absolute quantification (pg/mL) plus relative NPX. This format is useful in mechanistic studies, cell culture supernatants, or when validating findings from discovery panels.
Large-Scale & Population Studies
Although many studies are disease-adjacent, in non-clinical or translational biology, panels have been used in ageing, environmental exposure, or risk modelling. For instance, recent publication highlights show over 3000 peer-reviewed articles making use of PEA in proteomics / multiomics.
These studies illustrate that the Olink Inflammation Panel is not just for hypothesis generation but also for robust quantifiable profiling even in experimental settings.
Practical Tips: Designing Experiments & Avoiding Common Pitfalls
To make the most of the inflammation panel, here are good practices and common pitfalls to avoid:
Pre-study planning: Define precisely which proteins are of interest; this helps decide whether to use a standard panel or a custom Flex.
Replicates & randomisation: Include technical and biological replicates. Randomise sample order across plates to minimise batch or plate effects.
Control samples: Always include controls (e.g., untreated vs treated, baseline vs timepoints) with known expression levels. QC samples across batches enable estimation of variance.
Blanks / negative controls: To detect background or non-specific binding.
Handling missing / low values: Decide in advance how to treat proteins below LOD (e.g., imputing, excluding, flagging). Low abundance proteins may have more missingness.
Statistical correction: With many proteins, apply multiple testing corrections (e.g., FDR); consider effect sizes, not just p-values.
Metadata and sample annotation: Document sample collection time, freeze time, storage, subject metadata (age, sex, batch). These often explain unanticipated variation.
Emerging Trends & Innovations in Inflammation Proteomics with Olink
The field doesn't stand still—Olink and the larger proteomics community are pushing boundaries.
Higher-plex panels & explore platforms: Larger panels (e.g., Explore 384, 3072) allow broad system profiling, enabling discovery of novel inflammation networks.
Integration with other omics: Pairing protein data with transcriptomics, metabolomics, epigenomics for multi-layer insights into inflammation regulation.
Custom panels & absolute quantification: Tools like Olink Flex give both NPX and pg/mL values for select proteins. Useful in bridging discovery to mechanistic / translational biology.
Improved data analysis tools: Better normalization algorithms, batch correction, statistical models suited for multiplex proteome data. Public tools (e.g., BigOmics, OlinkAnalyze) are becoming more user-friendly.
Sample type expansion: Beyond plasma/serum — tissue lysates, supernatant from cell culture, organoids. Also better QC in low-volume, low-input setups.
If you want to deepen understanding in specific areas, the following internal articles are helpful:
"Exploring the Olink 96 and 48-Plex Panels: Key Differences" – good for detailed comparisons when selecting panel size.
"Olink Proteomics in Clinical Research: A Game Changer" – although clinical context, many principles about translation and high-throughput profiling apply in research settings.
Conclusion & Call to Action
The Olink inflammation panel offers powerful, multiplexed, sensitive profiling of inflammation mediators, with small sample requirements and strong reproducibility. For researchers in universities, R&D labs, and academic institutes, it enables mapping immune signalling and regulatory pathways with more depth than single-protein assays.
If you plan a study involving multiple inflammation‐related proteins, consider reaching out to us to:
- Discuss which Olink panel version (Target, Explore, Flex) suits your hypothesis
- Review your experimental design to optimize for low-abundance detection
- Get a quote for services, or request sample pilot runs with your specimens
Let's collaborate to turn your inflammation biology questions into quantifiable insights.
References
- Struglics A, Larsson S, Lohmander LS, Swärd P. Technical performance of a proximity extension assay inflammation biomarker panel with synovial fluid. Osteoarthr Cartil Open. 2022 Jul 7;4(3):100293. doi: 10.1016/j.ocarto.2022.100293. PMID: 36474941; PMCID: PMC9718077.
- Multiplex analysis of inflammatory proteins: A comparative study across multiple platforms
- Diniz, B.S., Chen, Z., Steffens, D.C. et al. Proteogenomic signature of Alzheimer's disease and related dementia risk in individuals with major depressive disorder. Nat. Mental Health 3, 879–888 (2025).
- Botey-Bataller, J., van Unen, N., Blaauw, M. et al. Genetic and molecular landscape of comorbidities in people living with HIV. Nat Med (2025).
- Liu, X., Zhuang, C., Liu, L. et al. Exploratory phase II trial of an anti-PD-1 antibody camrelizumab combined with a VEGFR-2 inhibitor apatinib and chemotherapy as a neoadjuvant therapy for triple-negative breast cancer (NeoPanDa03): efficacy, safety and biomarker analysis. Sig Transduct Target Ther 10, 237 (2025).








