Choosing the right panel matters more than choosing the platform when your goal is high-throughput immune profiling. For an immunology program with N≈800 plasma samples (plus a CSF subset), ≤20 µL available per subject, a cytokine-poor profile, a two-phase explore→validate plan, RNA-seq integration, and a four‑month milestone for first results, the pivotal decision is whether to start broad with Explore 3072/1536 and then confirm on Target 96 (Inflammation or Immuno‑Oncology) and/or Target 48 Cytokine. This guide compares these options with a focus on downstream validation continuity and multi‑omics integration—while staying neutral and evidence‑led.
If you're new to PEA/NPX and Olink workflows, see the background primer in Olink proteomics on PEA technology and research-ready data.
Key takeaways
- Prioritize continuity: Plan from day one how discoveries on Explore will bridge to Target 96/48 and orthogonal assays (e.g., ELISA/MS) and how protein signals will align with RNA‑seq pathways.
- Expect detectability trade-offs: Very high‑plex discovery may show higher below‑LOD for ultra‑low cytokines in cytokine-poor cohorts; use pilot plates and consider Target 48 Cytokine for absolute quant or improved low‑end capture.
- Design the workflow around constraints: ≤20 µL input, multi‑center batches, plate bridging, and a four‑month timeline all influence panel choice and batching strategy.
- Cost scales with strategy, not just plex: Per‑sample economics depend on batch design, bridge samples, re-runs, and validation phase planning—not just list price.
- There's no single "best" panel: Map panels to study phase and biological questions; broad discovery on Explore, focused confirmation on Target 96/48.
What "high‑throughput" means in Olink proteomics
In Olink proteomics, high‑throughput refers to measuring large protein sets efficiently using PEA (Proximity Extension Assay) with either qPCR (Target series) or NGS readouts (Explore series). Explore 3072 aggregates eight Explore 384 subpanels to cover roughly 2,900–3,072 proteins with PEA+NGS and typical inputs of 1–6 µL per sample. Explore 1536 uses four Explore 384 subpanels (~1,500 proteins); public, dedicated datasheets for Explore 1536 are limited—treat specs as indicative and verify per project on Olink's current documentation.
The Target 96 family measures 92 proteins per panel via PEA with ~1 µL input and a qPCR readout. For immune profiling, Target 96 Inflammation focuses on cytokines/chemokines/regulators, while Target 96 Immuno‑Oncology (used here as the closest proxy to "Immune Response") emphasizes checkpoints and tumor‑immune biology. Olink does not currently list a 96‑plex cytokine panel; the closest is Target 48 Cytokine (~45 cytokines), which also supports absolute quantification with calibrators.
For a deeper overview of Olink high‑throughput workflows and Explore scale, see the high‑throughput proteomics guide.
Comparison dimension 1 — Protein coverage and immune pathway breadth
- Explore 3072: The broadest coverage, spanning inflammation, cytokines/chemokines, complement, innate and adaptive immune regulators, and checkpoint‑related proteins. Best suited for hypothesis‑free discovery, pathway mapping, and network inference across large cohorts.
- Explore 1536: Mid‑scale breadth when you want substantial immune coverage but fewer subpanels. Often chosen when prior hypotheses narrow the search space or when input/time constraints favor fewer plates.
- Target 96 Inflammation: Focused coverage of the inflammatory axis—common cytokines/chemokines and regulatory proteins, enabling targeted hypothesis testing and confirmation.
- Target 96 Immuno‑Oncology: Immune checkpoints and tumor‑immune interactions; useful when discoveries point to checkpoint signaling or IO‑relevant mediators.
- Target 48 Cytokine (related): Concentrated on cytokines with an option for absolute quant. Valuable when cytokine signals are central to validation, particularly in cytokine‑poor matrices.
Think of coverage like a wide‑angle lens vs a zoom: Explore panels help you see the full landscape; Target panels let you zoom into the most relevant features once you know where to look.
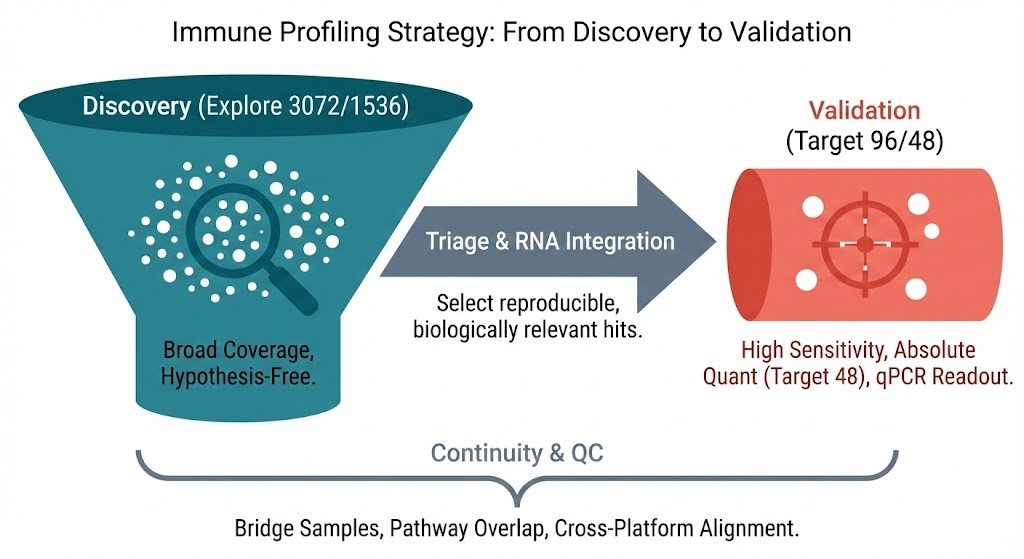 Strategic workflow for high-throughput immune profiling.
Strategic workflow for high-throughput immune profiling.
Comparison dimension 2 — Sensitivity and detectability for low‑abundance cytokines
Direct, analyte‑level head‑to‑head comparisons of Explore 3072/1536 vs Target 96/48 for immune cytokines and checkpoints are scarce in 2024–2026 public literature. Cross‑platform studies offer context:
- A 2024 comparison of Olink Target 96 with other multiplex technologies reported mid‑tier detectability overall, with some ultra‑low cytokines (e.g., IL‑13, IL‑4) showing lower detection rates, underlining the importance of matrix‑specific performance and per‑analyte variability, per Abe et al. (2024).
- Another 2024 analysis found Olink Target 48 Cytokine often more sensitive than some MSD V‑PLEX assays for shared cytokines but below MSD S‑PLEX for the ultra‑low end, reinforcing the need for analyte‑specific planning in cytokine‑poor cohorts, per McKinski et al. (2024).
Practical guidance:
- Pilot plates in your matrix (plasma ± CSF) are essential to estimate below‑LOD rates before committing to full runs.
- Where Explore discovery identifies cytokine signatures with marginal detectability, confirm on Target 48 Cytokine for absolute quant or bring in ultra‑sensitive orthogonal assays for specific analytes.
- Plan overlap between discovery and validation panels to avoid re‑targeting from scratch.
For cytokine‑specific panel considerations, see the Target 96 vs 48 plex explainer and the cytokine panel guidance.
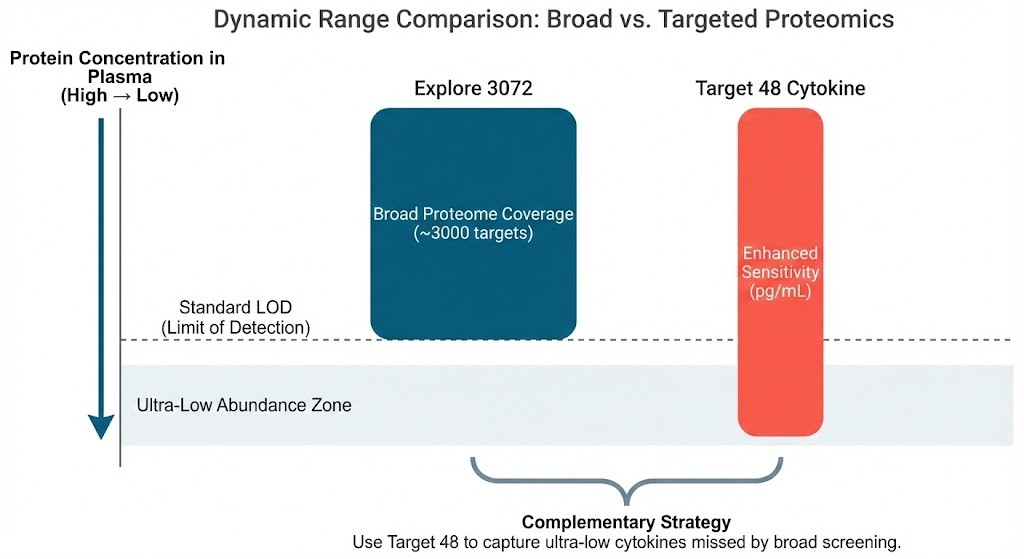 Complementary dynamic ranges of Explore and Target panels.
Complementary dynamic ranges of Explore and Target panels.
Comparison dimension 3 — Sample input, workflow, and operational practicality
- Sample input: Explore typically accommodates 1–6 µL depending on subpanel combinations; Target 96 uses ~1 µL per sample; Target 48 Cytokine similarly low inputs with calibrators for absolute quant.
- Plate format and bridging: Olink plates commonly run ~88 biological samples plus controls per 96‑well plate. Large cohorts benefit from identical biological bridge samples across plates to enable normalization and cross‑batch comparability—documented in the UK Biobank Explore 3072 plate layout FAQ and Olink manuals.
- Multi‑center robustness: Randomize subjects across plates and centers; model plate/run/lot/site as covariates; inspect QC plots (PCA/UMAP/boxplots) before downstream statistics.
- Timeline planning: Reserve time for pilot runs to de‑risk detectability and normalization; then scale with parallel batches aligned to the four‑month milestone.
Evidence:
- The UK Biobank pilot measured ~3,000 proteins in >54,000 participants using Explore 3072; plate layout and controls are detailed in the UKB Explore 3072 FAQ (2025).
- General Explore HT QC/normalization principles appear in the Olink Explore HT user manual.
- Practical normalization and bridging steps are summarized in this data analysis explainer.
Comparison dimension 4 — Cost structure and scalability for N≈800
Rather than quoting prices, consider cost drivers that materially affect per‑sample economics and total time‑to‑result:
- Batch architecture: Number of plates, parallelization, and bridge sample allocation.
- Detectability risk management: Pilot runs reduce re‑runs and downstream rework.
- Validation continuity: Selecting validation panels with high analyte overlap avoids repeating discovery on new targets.
- Multi‑center logistics: Shipping, scheduling, and QC coordination add real overhead.
- Analysis pipeline: Normalization, QC review, and integrated RNA‑seq analyses consume meaningful person‑time.
For the representative scenario, a well‑planned Explore discovery followed by targeted validation can meet a four‑month first‑results goal if pilot plates are front‑loaded, batches are parallelized, and QC sign‑offs are staged.
Priority: Continuity to downstream validation and RNA‑seq integration
Continuity is the primary decision axis. Here's how to design for it:
- Panel overlap: Prioritize analytes that appear on Explore and your intended Target 96 (Inflammation/Immuno‑Oncology) or Target 48 Cytokine, so hits translate without re‑targeting.
- Orthogonal confirmation: For critical biomarkers, plan ELISA or targeted LC‑MS confirmation. Explore vs MS comparisons support complementary strengths—use orthogonal assays for absolute quant and mechanism building, as discussed in Olink vs Mass Spectrometry.
- RNA‑seq integration: Align NPX signals with transcript clusters via correlation, pathway enrichment, and ligand–receptor analysis. Ensure consistent identifiers and metadata across omics. See multi‑omics integration guidance.
- Bridging strategy: Include overlapping biological bridge samples on every plate; assess alignment statistically and visually before progressing.
Side‑by‑side comparison table
| Panel | Proteins | Readout | Typical input | Immune focus notes | Continuity for validation |
| Explore 3072 | ~2,900–3,072 | PEA + NGS | 1–6 µL | Broad immune pathway coverage; discovery across cytokines, chemokines, checkpoints | Bridge to Target 96/48; overlap assays for validation; consider orthogonal MS |
| Explore 1536 | ~1,500 (indicative) | PEA + NGS | Low µL (verify) | Mid‑scale breadth; useful when narrowing hypotheses | Similar; confirm overlap with intended Target 96/48 |
| Target 96 Inflammation | 92 | PEA (qPCR) | ~1 µL | Focused inflammatory mediators | Strong for confirmation; high overlap with common inflammatory markers |
| Target 96 Immuno‑Oncology | 92 | PEA (qPCR) | ~1 µL | Checkpoints/tumor‑immune biology | Confirmation in IO contexts; checkpoint‑focused follow‑up |
| Target 48 Cytokine (related) | ~45 | PEA (qPCR) | ~1 µL | Cytokine‑centric; absolute quant available | Ideal for cytokine‑poor cohorts and absolute quant needs |
Notes: Explore 1536 public specs are limited; verify current inputs and coverage with Olink. Confirm matrix compatibility and panel combinations per project.
Which panel fits which immune profiling scenario?
- Early discovery in large cohorts (N≈800, cytokine‑poor plasma ± CSF): Explore 3072 if breadth is paramount and you need extensive pathway discovery; Explore 1536 when you want significant immune coverage with fewer subpanels.
- Hypothesis‑driven validation of inflammatory signals: Target 96 Inflammation to confirm specific cytokine/chemokine/regulatory hits emerging from Explore.
- Checkpoint‑centric or tumor‑immune follow‑ups: Target 96 Immuno‑Oncology to test checkpoint signaling hypotheses detected during discovery.
- Cytokine‑focused confirmation and quantitation: Target 48 Cytokine when low‑end detectability and absolute quant (pg/mL) are required.
- Longitudinal immune dynamics: Use Explore for initial pathway mapping, then Target 96/48 for consistent, cost‑efficient follow‑up across time points.
Common pitfalls when choosing high‑throughput immune panels
- Over‑valuing protein count while ignoring pathway redundancy and analyte overlap with validation panels.
- Locking into a panel too early during exploration, constraining downstream validation options.
- Underestimating detectability limits in cytokine‑poor matrices and skipping pilot plates.
- Neglecting plate bridging and multi‑center randomization, leading to batch effects and re‑runs.
FAQ: High‑throughput Olink panels for immune profiling
What's the difference between Explore and Target panels for immune profiling?
Explore uses PEA with NGS readouts to cover thousands of proteins for discovery; Target uses PEA with qPCR to measure ~92 proteins per panel for focused validation. Explore is breadth; Target is precision.
Is Target 96 sufficient for large immune cohort studies?
Yes for validation or hypothesis‑driven studies. For initial discovery in large, complex cohorts, Explore provides broader coverage to identify the most promising targets.
How does panel choice affect detectability of cytokines?
In cytokine‑poor cohorts, very high‑plex discovery may produce higher below‑LOD fractions for ultra‑low cytokines. Target 48 Cytokine or ultra‑sensitive orthogonal assays may improve confidence for specific analytes.
Which panel is better for exploratory vs validation‑stage immune research?
Explore 3072/1536 for exploratory breadth; Target 96 (Inflammation or Immuno‑Oncology) and Target 48 Cytokine for validation and quantitation.
Can different panels be combined across project phases?
Yes—and that's often optimal. Design overlap between Explore hits and your chosen Target panels; include bridge samples on every plate to ensure continuity.
Conclusion — Choose panels based on study goals, not just throughput
There's no universally "best" panel in Olink proteomics. For N≈800, cytokine‑poor plasma ± CSF cohorts, an Explore‑first strategy followed by Target 96/48 validation typically balances discovery breadth, detectability needs, and operational realities. The winning move is to prioritize continuity: overlap analytes across phases, plan bridge samples, and align NPX signals with RNA‑seq mechanisms. Do that, and your four‑month milestone is far more likely to deliver results that are not only fast, but decision‑ready.




