- Panel Features
- Panels List
- Workflow
- Why Creative Proteomics
- Demo
- Sample Requirements
- Case
- FAQ
- Sample Submission Pack
Customized and Ready-to-Use Olink Proteomics Panel for Aging Diseases
Why Analyze Aging Diseases?
1. Longitudinal Biological Monitoring – Enables the study of progressive, time-dependent changes in protein expression and pathway activity associated with physiological decline.
2. Systemic Insight into Age-Related Processes – Captures organism-wide molecular shifts in metabolism, inflammation, and cellular communication that underlie the aging process.
3. Dynamic Biomarker Discovery – Identifies circulating proteins and secretory factors reflecting cellular senescence, mitochondrial dysfunction, and stress response mechanisms.
By leveraging proteomic analysis in aging research, scientists gain a comprehensive view of molecular aging mechanisms, accelerating the discovery of biomarkers for biological age and resilience.
What We Provide
- Aging biology–focused programs — Cellular senescence/SASP, mitochondrial dysfunction, nutrient sensing (mTOR/AMPK/IGF), proteostasis, inflammaging, DNA damage/repair.
- Study design for aging cohorts — Cross-sectional and longitudinal frameworks, visit schedules, randomization, and batch-bridging controls for multi-timepoint comparability.
- Custom panels & protein signatures — 21–384+ plex selection; pathway modules for senescence/stress/longevity; composite indices and protein aging clocks development and benchmarking.
- Sample & model coverage — Plasma/serum and other biofluids (e.g., urine/CSF); support for human cohorts and preclinical models (mouse/rat/NHP) with cross-species ortholog mapping.
- Olink + multi-omics integration — Combine proteomics with bulk/single-cell/spatial RNA-seq, epigenomics, and metabolomics for pathway/network analyses and subtype discovery.
- Biostatistics & reporting — NPX matrix with full QC; age-association and trajectory (mixed-effects) modeling, enrichment/co-expression networks; publication-ready figures and optional interactive dashboards.
- Orthogonal verification — ELISA/MSD, targeted LC-MS, and replication across cohorts/platforms to strengthen research conclusions.
This service supports:
- Identification of novel aging biomarkers
- In-depth mechanistic studies of aging processes
- Analysis of inflammatory and metabolic changes in aging models
- Scalable solutions for proteomic aging research
Features of the panel
- Species: Optimized for human proteome profiling in aging studies.
- Design: Customizable target selection via bioinformatics interface for age-related pathways.
- Proteins: 21- to 384-plex panels targeting senescence-associated secretions, stress response markers, and longevity pathways.
- Sample: 1 µL plasma/serum per analysis for minimal sample consumption.
- Readout: NPX-based quantification for normalized protein expression values.
- Platform: PEA-based detection system ensuring high sensitivity and reproducibility.
- High Sensitivity: Detects low-abundance proteins (to pg/mL level), crucial for aging biomarkers like inflammatory cytokines and metabolic regulators
- High Reproducibility: Maintains inter-assay CV <10% for reliable data across aging study timepoints.
- Tailored Panel Design: Pre-configured or customizable panels focusing on senescence, stress response, longevity, and metabolic pathways.
List of Our Aging Proteome Assay Panel
Protein category
The Aging Proteome Assay Panel quantitatively analyzes proteins across six core functional categories essential for aging biology research: Metabolic Regulators, Senescence-Associated Secretory Proteins, Stress Response Mediators, DNA Repair Factors, Protein Homeostasis Components, and Intercellular Signaling Molecules (see Table: List of Aging Biomarker Assay).
This panel encompasses fundamental biological processes associated with aging, including cellular senescence, mitochondrial dysfunction, nutrient sensing, proteostasis maintenance, and inflammatory regulation, featuring strategically selected targets implicated in longevity pathways and age-related physiological decline. Through systematic literature curation and experimental validation, we incorporate rigorously vetted biomarkers associated with telomere maintenance, oxidative damage response, autophagy, and epigenetic regulation, providing researchers with a specialized tool for comprehensive proteome investigations in diverse aging research areas, including longevity studies, age-related functional decline, and intervention mechanism research. The panel's optimized design enables focused examination of aging-related protein interaction networks while preserving the analytical depth required for mechanistic discovery in aging biology.
Table. List of Aging Biomarker Assays
Protein Functions
Biological process
primarily involved in key biological processes, including cellular senescence regulation, protein homeostasis maintenance, mitochondrial function modulation, DNA damage response pathways, and metabolic adaptation mechanisms, providing researchers with comprehensive insights into fundamental aging mechanisms and longevity-associated biological networks.
Disease area
primarily utilized in research areas involving cellular senescence models, age-related metabolic dysfunction studies, chronic low-grade inflammation research, mitochondrial dysfunction mechanisms, and protein aggregation studies, enabling investigators to explore fundamental biological mechanisms underlying aging processes and develop interventions targeting age-related physiological decline.
The Application of Aging Proteome Assay.
The Aging Proteome Assay enables high-throughput quantification of protein biomarkers across biological pathways, providing researchers with a powerful tool for:
- Discovery of protein networks underlying cellular senescence and longevity mechanisms in model organisms;
- Mechanistic investigation of mitochondrial dysfunction and oxidative stress responses in aging studies.
- Identification of molecular signatures associated with proteostasis decline and genomic instability in aging research.
- Analysis of nutrient-sensing pathway alterations and metabolic adaptations in age-related studies.
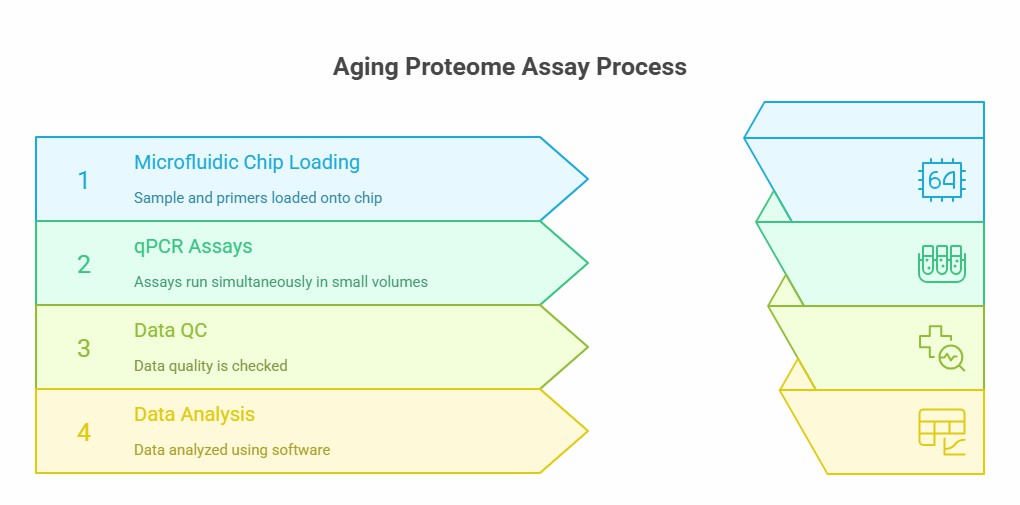
Workflow of Olink Proteomics
Why Creativ Proteomics
Aging-Specific Analytical Frameworks
Preconfigured workflows for cellular senescence, mitochondrial dysfunction, and proteostasis pathways with 50+ validated biomarker ratios.
Multi-Omics Integration
Compatible with transcriptomic, epigenomic, and metabolomic data for systems-level aging biology research.
Longitudinal Study Optimization
Stable normalization controls and batch correction algorithms are designed for long-term aging studies.
Pathway-Centric Analysis
Automated bioinformatics tools for aging pathway enrichment (e.g., mTOR, AMPK, DNA damage response).
Demo Results of Olink Data
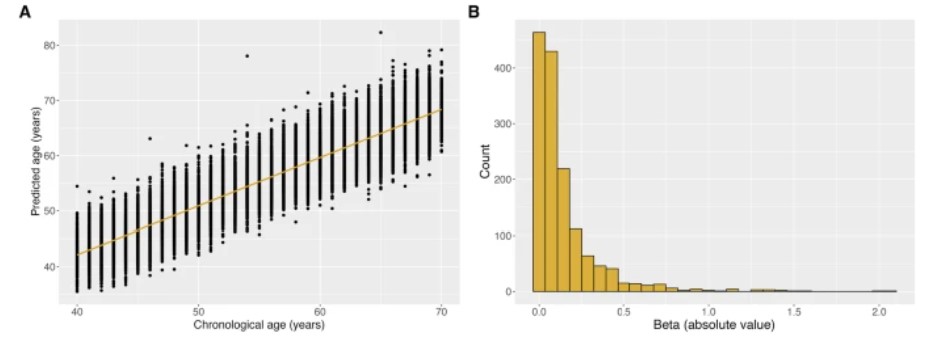 Figure 1: Performance and characteristic contributions of aging clock models. (Mörseburg, A., et al. 2025)
Figure 1: Performance and characteristic contributions of aging clock models. (Mörseburg, A., et al. 2025)
Sample Requirements
| Sample Type | Recommended Sample Size | Sample Quality | Pre-treatment and Storage | Sample Transport |
| Plasma/Serum/Body Fluid | The minimum input volume per assay is 40 µL. | For optimal results, maintain concentrations between 0.5 and 1 mg/mL. | For long-term preservation, aliquot samples and maintain at -80°C. | Ship sealed samples on dry ice (-80°C). |
| Tissue | ||||
| Cells | ||||
| Exosomes | ||||
| Other |
Case Study

Healthy Aging Metabolomic and Proteomic Signatures Across Multiple Physiological Compartments
Journal: Aging cell,
Year: 2025
- Background
- Methods
- Results
The study of biomarkers in biological fluids and tissues has expanded researchers' understanding of the biological processes that drive the physiological and functional manifestations of aging. However, most of these studies are limited to one biological part, a practice that fails to recognize that aging affects the entire body in general. Modeling hundreds of metabolites and proteins at multiple sites simultaneously may provide a more detailed picture of healthy aging and reveal differences between temporal and biological aging. Here, this study reports proteomic analysis of plasma and urine samples from healthy men and women aged 22 to 92 years.
Detection of proteins is performed by Olink Explore [Type 1536] according to the manufacturer's instructions. The technology behind the Olink protocol is based on proximity extension assay (PEA) and reads by next-generation sequencing (NGS). The assay is capable of detecting up to 1536 proteins in 90 samples simultaneously using only 2.8 microliters of serum/plasma. In short, oligonucleotide-labeled antibody probes designed for each protein bind to their targets, bringing complementary oligonucleotides close together and allowing them to hybridize. The addition of DNA polymerase results in the extension of hybrid oligonucleotides, resulting in unique protein recognition "barcodes". Next, library preparation adds the sample identification index and the nucleotides needed for Illumina sequencing. Before sequencing with the Illumina NovaSeq 6000, libraries undergo a bead-based purification step and are evaluated for quality using an Agilent 2100 bioanalyzer. Raw count data were generated using bcl2counts (v2.2.0) and were quality controlled, standardized, and converted to Olink's unique unit of relative abundance, "normalized protein expression" (NPX). The standardization of data is carried out through internal extension control and external board control to adjust for differences in internal and external operations. All test validation data is available on the manufacturer's website.
Plasma and urine proteomics studies were performed in healthy men and women (from the GESTALT study with an age range of 22 to 92 years) using the Explore 1536 Olink (Proximity Extension Analysis (PEA)) technology, at baseline (n = 101; Figure 1a) and at 2 years of follow-up (n = 65), respectively. At the start of the GESTALT study, none of the participants had chronic disease, including cardiovascular, pulmonary, gastrointestinal, autoimmune, and metabolic disease, did not smoke, and had no active cancer within the past 10 years. In this study, the investigators performed a cross-sectional analysis of measured proteins in plasma and urine at baseline and 2-year follow-up. At baseline, relative concentrations of 1432 and 1055 proteins were detected, respectively. There was a clear separation of proteins in plasma and urine between different sites, with the first three principal components explaining the 52.4% variance between the two parts at baseline (Figure 1b). Linear regression models adjusted for sex, ethnicity, and body mass index were used to identify proteins that were significantly associated with age at baseline (Figure 1c, d) and at two-year follow-up (Figure 1c, d). As shown in Figure 1c, most age-related protein levels in plasma increased (increasing with age) while most age-related protein levels in urine decreased (decreased with age), positively correlated with age (Figure 1d).
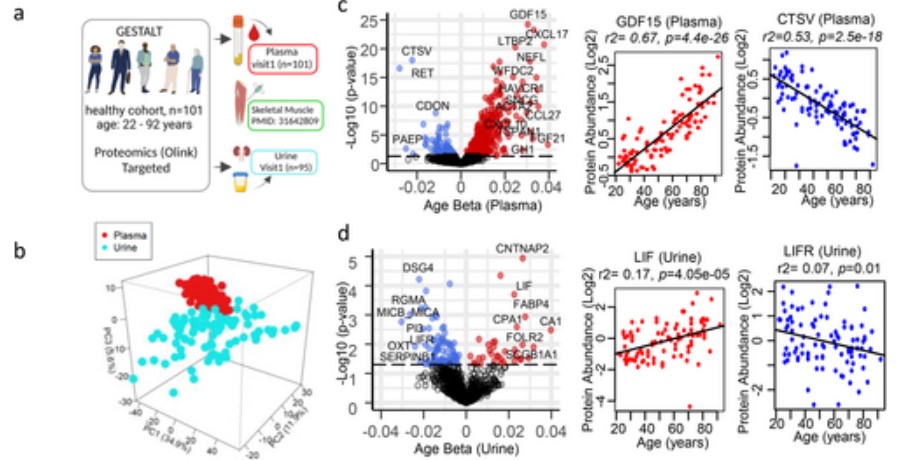 Figure 1. Analysis of the aging process in plasma and urine in a healthy person. ( Moaddel, R., et al. 2025)
Figure 1. Analysis of the aging process in plasma and urine in a healthy person. ( Moaddel, R., et al. 2025)
FAQs
What research applications does this assay support?
- Investigation of cellular senescence and proteostasis mechanisms
- Longitudinal monitoring of age-related protein dynamics in model organisms
- Nutrient-sensing pathway analysis (e.g., mTOR, AMPK signaling)
- Mitochondrial dysfunction and oxidative stress studies
Can researchers customize protein targets?
Yes, the assay allows selection of 20–500 analytes from a validated library of 1,200+ aging-related proteins, with options to include pathway-specific modules (e.g., autophagy, DNA damage response) while maintaining sensitivity and reproducibility standards.
How are data analyzed and delivered?
Processed data includes normalized protein expression (NPX) values, quality control metrics, and automated pathway enrichment analysis for aging-related biological processes (e.g., telomere maintenance, protein aggregation).
Does the platform support multi-omics integration?
Yes, the assay provides seamless integration with transcriptomic and metabolomic data through unified bioinformatics pipelines, enabling systems-level analysis of aging mechanisms.
References
- Mörseburg, A., Zhao, Y., Kentistou, K. A., et al. (2025). Genetic determinants of proteomic aging. npj aging, 11(1), 30. https://doi.org/10.1038/s41514-025-00205-4
- Moaddel, R., Candia, J., Ubaida-Mohien, C., et al. (2025). Healthy Aging Metabolomic and Proteomic Signatures Across Multiple Physiological Compartments. Aging cell, 24(6), e70014. https://doi.org/10.1111/acel.70014