- Panel Features
- Panels List
- Workflow
- Why Creative Proteomics
- Demo
- Sample Requirements
- Case
- FAQ
- Sample Submission Pack
Customized and Ready-to-Use Olink Proteomics Panel for Tissue Sample
Why Analyze Tissue Samples?
1. Spatial Biological Context – Tissue samples provide precise protein localization within anatomical structures, enabling the study of cellular microenvironment interactions and regional molecular distributions.
2. Direct Pathway Investigation – Enables focused analysis of cell-specific signaling cascades, post-translational modifications, and functional protein complexes directly at their source.
3. Comprehensive Molecular Profiling – Captures tissue-specific proteins, structural components, and low-abundance targets often diluted or absent in circulating biofluids.
By leveraging tissue proteomics, researchers gain critical insights into localized biological mechanisms, accelerating the discovery of context-specific biomarkers for fundamental mechanistic research.
What We Provide
Our Olink PEA platform delivers high-sensitivity, high-throughput protein quantification.
- Disease- & tissue-focused programs — Oncology/TME, brain, fibrosis, skin & gut inflammation, metabolic liver/kidney; supports FFPE and fresh-frozen (incl. TMA).
- Multi-species & model coverage — Human cohorts, mouse/rat, PDX, organoids; cross-species ortholog mapping and signature transfer.
- Custom panels & study design — 21–384+ plex target selection, hypothesis matrices, randomization and batch-bridging controls; pilot feasibility studies.
- Spatial & pathology integration — ROI-guided sampling, macro/laser microdissection (LCM), H&E/IHC/IF co-localization; pathology review support.
- Olink + multi-omics integration — Bulk/single-cell/spatial RNA-seq, WES/WGS, metabolomics; pathway/network analysis and subtype stratification.
- Biostats, reporting & validation — NPX matrix + QC, differential/longitudinal models, enrichment analyses; ELISA/MSD, targeted LC-MS, IHC/qPCR for orthogonal confirmation.
This service is optimized for:
- Tissue-specific biomarker discovery and validation
- Tumor microenvironment and immune cell profiling
- Mechanistic investigations in oncology, neuroscience, and inflammation research
- Scalable, ready-to-use workflows designed for tissue proteomic studies
Features of the panel
- Species: Human proteome optimized
- Design: Custom panel selection via bioinformatics interface.
- Proteins: Supports 21- to 384+-plex panels targeting cytokines, immune responses, and specific pathways.
- Sample: 1-6 µL Tissue samples per analysis.
- Readout: NPX quantitative readout
- Platform: PEA-based detection platform.
List of Our Tissue Proteome Assay Panel
Protein category
The Tissue Proteome Assay Panel quantitatively analyzes proteins across many essential functional categories for biological research: Metabolic Regulators, Signaling Transducers, Immune Modulators, Structural Matrix Proteins, Hormones and Binding Proteins, and Cellular Stress Mediators (see Table: List of Tissue Biomarker Assay).
This panel encompasses fundamental biological processes, including intercellular communication, metabolic adaptation, inflammatory regulation, and tissue reorganization, featuring precisely selected targets implicated in core physiological pathways. Through systematic literature curation, we integrate rigorously validated biomarkers associated with tissue-specific functions, cellular microenvironment interactions, and architectural remodeling, providing researchers with a comprehensive platform for spatially resolved proteome investigations across diverse research areas, including developmental biology, cellular adaptation studies, and microenvironmental response mechanisms. The panel's organized structure facilitates focused examination of protein interaction networks while preserving the analytical depth necessary for mechanistic research in tissue biology.
Table. List of Tissue Biomarker Assays
Protein Functions
Biological process
The Tissue Proteome Assay enables comprehensive investigation of fundamental biological processes within complex tissue architectures, including the regulation of metabolic zonation patterns across tissue compartments, spatiotemporal dynamics of intracellular signaling networks in response to localized stimuli, adaptive stress response mechanisms in pathological or experimental conditions, coordinated immune cell interactions within tissue microenvironments, and the maintenance of structural and functional homeostasis through extracellular matrix remodeling and cell-cell communication.
Disease area
The Tissue Proteome Assay provides a powerful platform for investigating disease mechanisms within native tissue contexts, enabling researchers to study pathological processes such as dysregulated metabolic pathways in metabolic disorders, aberrant signaling cascades in cancer microenvironments, compromised cellular stress responses in neurodegenerative conditions, disrupted immune coordination in inflammatory diseases, and loss of tissue homeostasis in fibrotic and degenerative conditions.
The Application of Tissue Proteome Assay.
The Tissue Proteome Assay enables spatially resolved, high-fidelity quantification of protein expression and modifications in complex tissue architectures, providing researchers with an advanced tool for:
- Systematic mapping of tumor microenvironment heterogeneity and immune cell infiltration in cancer biology models.
- Mechanistic investigation of neuroinflammatory pathways and neuronal signaling protein networks in brain tissue research;
- Comprehensive analysis of extracellular matrix remodeling in fibrotic diseases and engineered tissue constructs;
- Discovery of protein signatures associated with metabolic zonation in organ-specific studies (e.g., liver lobules, kidney nephrons);
- Spatially resolved profiling of cell-cell communication networks and ligand-receptor interactions in tissue contexts;
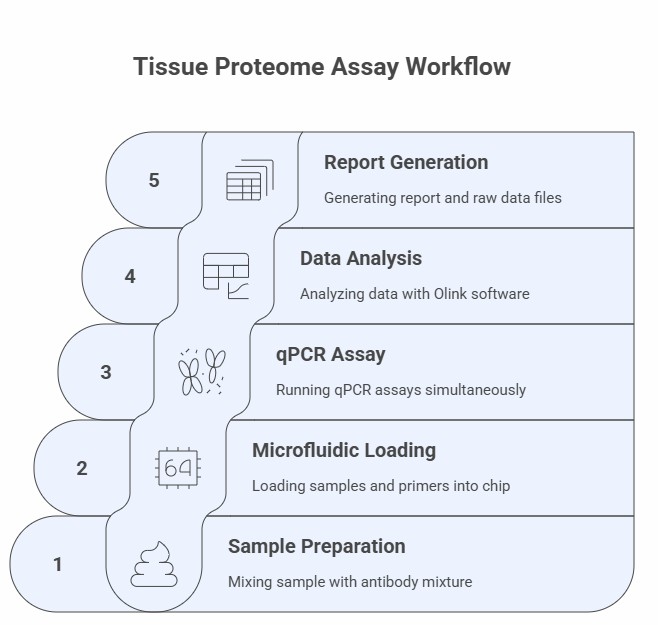
Workflow of Olink Proteomics
Why Creativ Proteomics
Comprehensive Tissue Matrix Validation
The panels are rigorously validated for diverse tissue types, including fresh-frozen biopsies, FFPE sections, and organoids, with optimized lysis protocols provided for each.
Robust Performance in Complex Lysates
Proprietary normalization controls and PEA technology ensure high specificity and reproducibility, minimizing interference from complex tissue homogenates.
Flexible Panel Customization
Tailor panels from a validated library of 3,500+ proteins to focus on specific tissue pathways or experimental perturbations.
Advanced Data Visualization Tools
Utilize integrated software for spatial heatmaps, protein co-expression networks, and pathway enrichment analysis directly within tissue research contexts.
Demo Results of Olink Data
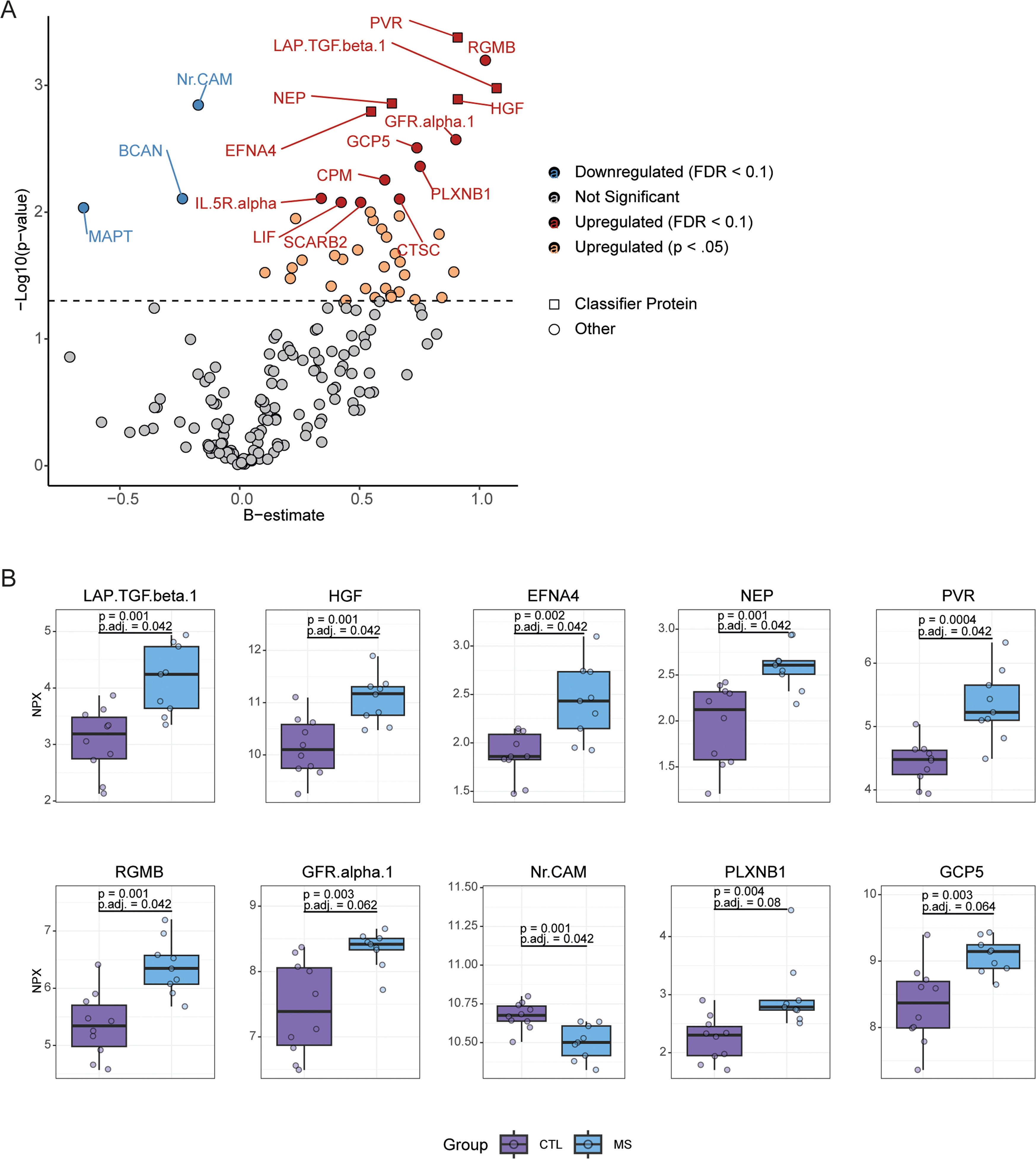 Figure 1: Altered expression of neuroinflammatory proteins in the cortical tissue of multiple sclerosis patients. (van der Stok, H., et al. 2025)
Figure 1: Altered expression of neuroinflammatory proteins in the cortical tissue of multiple sclerosis patients. (van der Stok, H., et al. 2025)
Sample Requirements
| Sample Type | Recommended Sample Size | Sample Quality | Pre-treatment and Storage | Sample Transport |
| Tissue | A 40 µL minimum volume is required per assay. | Optimal concentration range: 0.5-1 mg/mL. | For long-term storage, aliquot and preserve at -80°C. | Ship sealed specimens on dry ice (-80°C). |
Case Study
High content imaging shows distinct macrophage and dendritic cell phenotypes for psoriasis and atopic dermatitis
Journal: Scientific reports,
Year: 2025
- Background
- Methods
- Results
Atopic dermatitis (AD) and plaque psoriasis (PSO) are both multifactorial chronic inflammatory skin diseases with different immunological characteristics. In psoriasis, Th1, Th17, and Th22 T cell subsets play a pathogenic role with the involvement of multiple cytokines. Th17 (Tc17) type T cells are currently thought to be the main drivers of psoriasis. AD is associated with a type 2 immune response, and AD is often accompanied by increased IgE production and allergy or asthma symptoms. Notably, some types of AD have a significant IL-17 component and have histopathological features similar to those of psoriasis type 1. Thus, although psoriasis and AD share common features of chronic inflammation, their underlying immune processes involve different T cell responses, cytokine profiles, and immune cell activation modalities. Understanding these differences is crucial for developing targeted and effective treatments for each disease.
Tissue samples were lysed in 100 microliters of RIPA buffer (50 mmol of Tris-HCl, pH 7.4, 150 mmol of NaCl, 1 mmol of EDTA, addition of 1× protease inhibitor mixture). Sonicate samples in three 10-second sessions at 70% power using the SONOPULS HD2070 MS73 ultrasound. Centrifugation was subsequently performed (1300x gravity, 5 min), and the total protein concentration in the supernatant was determined using a Nano Drop UV/VIS spectrophotometer. All samples were adjusted to a final protein concentration of 1 mg/ml. Expression of inflammatory markers was determined on Biomark HD using the Olink Target 96 Inflammation Kit (as recommended by the manufacturer), and NPX™ labeling software was used for quality control and data processing. Data is presented as NPX values (any unit of Olink, represented by a log-2 scale).
To compare the immune response in patients with dermatomyositis (PSO) and rheumatoid arthritis (AD), researchers first measured the expression of 92 inflammatory markers using the Olink proteomics platform. In this study, skin biopsy samples from 14 patients with dermatomyositis and 14 patients with rheumatoid arthritis were compared. Thirteen biopsy samples from healthy donors were used as controls. Of the 92 proteins studied, 70 were detectable in at least one group, and 35 of the markers showed significant upregulation or downregulation in at least one group compared to healthy subjects. Principal component analysis differentiated samples from patients and healthy donors based on these 35 inflammatory markers, but could not distinguish between dermatomyositis and rheumatoid arthritis (Figure 1A). Dermatomyositis and rheumatoid arthritis modulate 14 inflammatory markers similarly, while 14 markers are specific to dermatomyositis and 7 are specific to rheumatoid arthritis (Figure 1B). The results showed that the expression of IL-17A, IFN-γ, TNF, VEGF-A, CCL7, and IL-12B, typical pro-inflammatory mediators that promote PSO progression, was significantly upregulated in the Pso group, while not in the AD group (Figure 1B). In AD biopsy samples, only 5 markers were specifically upregulated, including several mediators involved in cell differentiation and survival (RANKL, osteoprotegerin, leukemia inhibitory factor (LIF), and TRAIL) (Figure 1B). Importantly, among the AD-specific markers are the CCR1 ligand CCL13 and LIF, which can inhibit the pro-inflammatory response of macrophages and DC17. Taken together, these data highlight the different immune responses between AD and PSO, but also indicate difficulties in differentiating between the two diseases in principal component analysis, as there is significant overlap in the regulation of immune mediators.
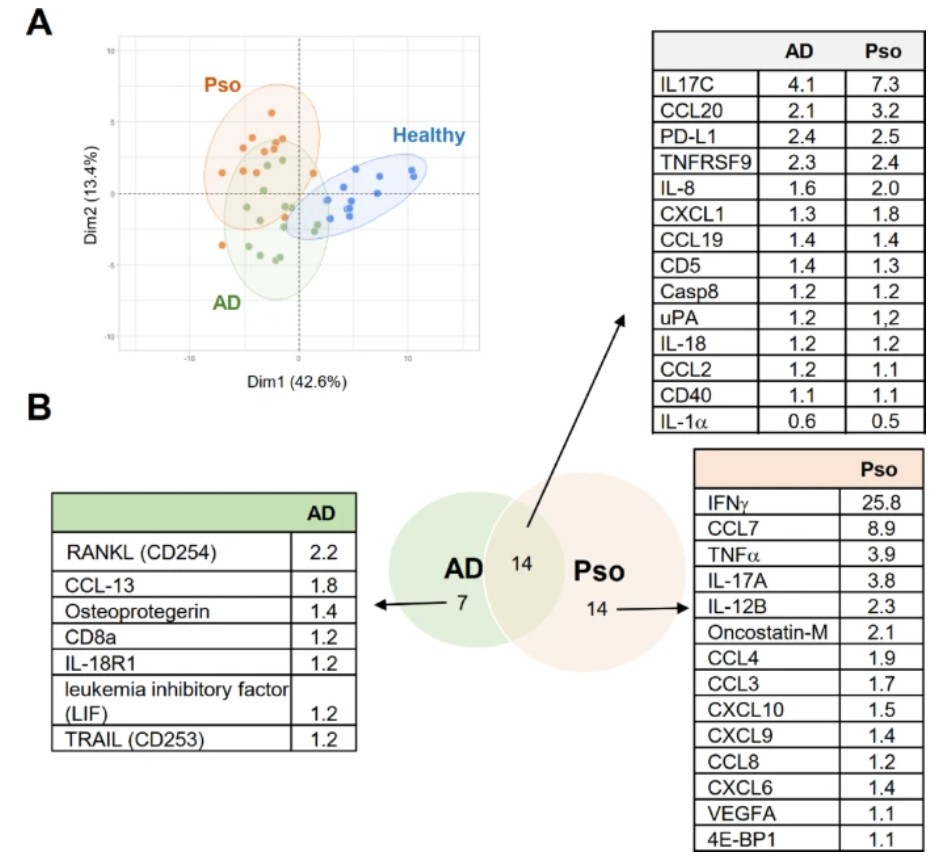 Figure 1. Analysis of differences in inflammatory marker levels in skin biopsy samples from patients with dermatoid disease and atopic dermatitis. ( Behr, N. J., et al. 2025)
Figure 1. Analysis of differences in inflammatory marker levels in skin biopsy samples from patients with dermatoid disease and atopic dermatitis. ( Behr, N. J., et al. 2025)
FAQs
What types of tissue samples are compatible with the Olink platform for proteomic analysis?
The technology is optimized for a variety of tissue types, including fresh-frozen biopsies, tissue explants, and patient-derived xenografts. The platform provides detailed protocols for tissue homogenization and protein extraction to ensure compatibility and maximize protein yield for downstream analysis.
How does the platform handle the complexity of tissue lysates compared to body fluids?
The assay incorporates advanced normalization controls and is specifically validated for the high-background matrix of tissue lysates. The PEA technology's dual antibody recognition ensures high specificity, minimizing non-specific binding and allowing for precise quantification of target proteins even in complex tissue homogenates.
Can we customize a protein panel specifically for our tissue-based research pathway?
Absolutely. Researchers can configure tailored panels from our validated library of over 3,500 human protein targets. This allows for the creation of focused panels for specific tissue pathways (e.g., tumor microenvironment, neuronal signaling) or broader panels for exploratory discovery, all while maintaining stringent sensitivity and reproducibility.
How does the tissue proteomic data integrate with other genomic data from the same samples?
The platform provides built-in compatibility for multi-omics integration. Data is delivered in unified file formats that are directly compatible with transcriptomic (RNA-seq) and genomic datasets. Specialized bioinformatics tools are included for cross-omics pathway mapping and co-expression network analysis, enabling a systems biology approach to tissue research.
References
- van der Stok, H., Coenen, L., Weil, M., et al. (2025). Proximity Extension Assay identifies new targets of grey matter pathology in multiple sclerosis. Multiple sclerosis and related disorders, 102, 106626. Advance online publication. https://doi.org/10.1016/j.msard.2025.106626
- Behr, N. J., Pierre, S., Ickelsheimer, T., et al. (2025). High content imaging shows distinct macrophage and dendritic cell phenotypes for psoriasis and atopic dermatitis. Scientific reports, 15(1), 18904. https://doi.org/10.1038/s41598-025-99727-w