- Overview
- Panels List
- Applications
- Workflow
- Why Creative Proteomics
- Demo
- Sample Requirements
- FAQ
- Sample Submission Pack
Why Integrate Olink Proteomics with miRNA?
The integration of Olink and miRNA hinges on dual-recognition mechanisms and functional genomics: miRNAs bind target mRNAs to suppress translation or trigger decay, directly influencing protein expression. Using Olink' s PEA technology for sensitive protein detection and miRNA methods (e.g., antagomirs, sponges, mimics) to block or restore miRNAs, this platform gives a full picture of gene networks and protein pathways in health and disease.
miRNA
1) Definition: miRNAs are endogenous, single-stranded, non-coding RNAs (~19–25 nucleotides) that regulate gene expression post-transcriptionally.
2) Biogenesis Pathway:
- Transcription: miRNA genes are transcribed by RNA polymerase II (Pol II) into primary transcripts (pri-miRNA), which are 5ʹ-capped and polyadenylated.
- Nuclear Processing: Drosha-DGCR8 complex cleaves pri-miRNA into pre-miRNA. Pre-miRNA is exported to the cytoplasm via Exportin-5 and RanGTP.
- Cytoplasmic Maturation: Dicer enzyme cleaves pre-miRNA into a short duplex (miRNA). Guide strand loads into the RNA-induced silencing complex (RISC), while the passenger strand is degraded.
3) Mechanism of Gene Silencing:
- Translational Repression: miRNA binds to the 3ʹ-UTRof target mRNA with imperfect complementarity. RISC blocks ribosome binding or translation initiation/elongation, suppressing protein synthesis without degrading mRNA.
- mRNA Degradation: Near-perfect complementarity between miRNA and mRNA triggers endonucleolytic cleavage by AGO2.
Olink Technology
Proximity Extension Assay (PEA)
1) Antibody pairs conjugate to DNA oligonucleotides.
2) Antibody pairs bind to target protein.
3) Upon co-localization, the DNA strands hybridize and extend into amplifiable templates.
4) Quantifiable DNA signals via qPCR or NGS.
Advantages
- Scalability and Sample Efficiency: Olink Explore HT profiles 5,400+ proteins per run; miRNA assays require minimal RNA input. Unified bioinformatics pipelines streamline multi-omics data fusion.
- Unprecedented Sensitivity and Specificity: Olink PEA detects low-abundance proteins in 1–6 µL of serum/plasma, overcoming matrix interference. miRNA tools exhibit high binding affinity and metabolic stability for in vivo efficacy.
- Multi-omics Integration: Correlates protein biomarkers (Olink) with miRNA regulators to identify master switches in disease pathways.
Olink & miRNA Platform: Selecting the Right Panel
Recommended Panels for Integrated Analysis
The choice of panels prioritizes disease relevance, pathway coverage, and scalability for miRNA-protein co-analysis. The panels specifically suitable for combining Olink proteomics with miRNA analysis are the Olink Explore 384 panels, Target 96 Immuno-Oncology/Cell Regulation/Oncology II/III panels, Target 48 Cytokine panels, and Customized Olink Flex panels. The reasons are as following:
- Explore 384 panels enable broad, hypothesis-free profiling across 384 high-value biomarkers in specific disease domains, capturing system-wide proteome changes to correlate with miRNA regulators.
- Target 96 panels provide focused, deep interrogation of disease-specific pathways with minimal sample consumption, essential for validating miRNA-mediated proteomic alterations identified in screening.
- Target 48 cytokine panels offer rapid, cost-effective quantification of signaling molecules critical for miRNA-regulated immune/inflammatory processes.
- Custom Olink Flex allows bespoke customization targeting miRNA-associated proteins, ensuring precise alignment with miRNA functional data.
Here is a list of recommended panels we offer:
Table. List of Olink Panels
Additional Options
In addition to the panels above, we also offer:
- Explore HT
- Explore 3072 (3072 proteins)
- Explore 1536 (1536 proteins)
- Target 96
- Custom Olink Flex systems
Our team can assist in designing an integrated Olink–miRNA strategy tailored to your project goals.
Applications
Biological Mechanism Exploration
- Researchers can map upstream regulatory networks to downstream protein expression changes. For instance, miRNA-targeted genes are cross-referenced with Olink-detected proteins to identify inverse expression patterns, revealing functional regulatory axes in cellular or tissue systems.
Multidimensional Biomarker Identification
- Combining miRNA and protein signatures enhances the robustness of biomarker panels. Olink's high-sensitivity protein quantification and miRNA profiling from biofluids enable the discovery of composite biomarkers. These panels improve predictive accuracy for biological condition assessment compared to single-molecule approaches.
Therapeutic Response Monitoring
- Dual miRNA-protein analysis tracks dynamic molecular shifts during interventions. For example, specific miRNAs and Olink-measured proteins can jointly reflect temporal changes in biological responses, providing granular insights into efficacy and mechanistic adaptations.
Combined RNA-Protein Therapeutics Development
- Co-delivery systems simultaneously transport miRNA modulators and protein-targeting agents. This strategy amplifies regulatory effects, such as suppressing oncogenic pathways or accelerating tissue repair, validated via Olink-measured protein pathway alterations.
Tissue Regeneration and Repair Optimization
- Synchronized miRNA inhibitors and protein-focused formulations enhance cellular repair processes. Metrics like migration rates, structural protein expression, and inflammation markers demonstrate synergistic improvements in tissue restoration models.
Workflow of Olink & miRNA
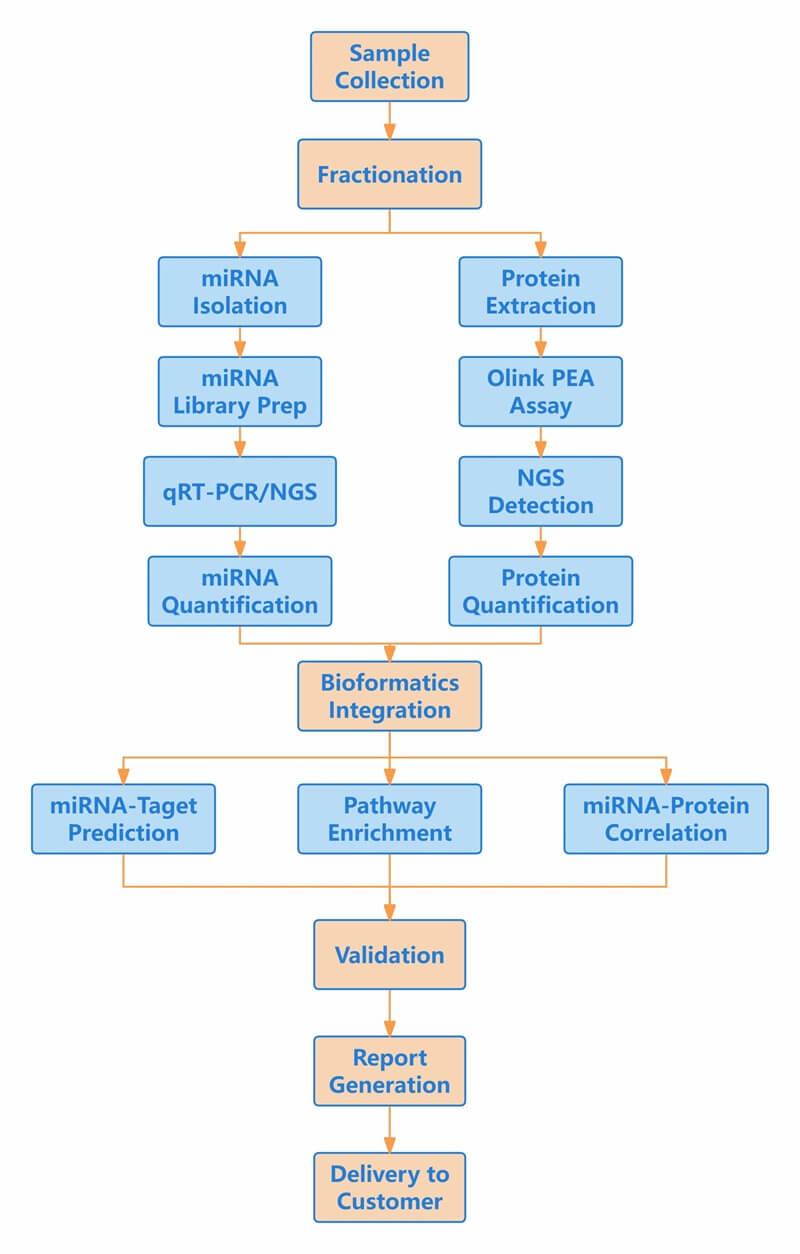 Figure 1: Workflow of integrated Olink proteomics and miRNA.
Figure 1: Workflow of integrated Olink proteomics and miRNA.
Why Creativ Proteomics
Multi-Omics Data Integration and Bioinformatics
- Unified Data Analysis: Bioinformatics pipelines to correlate miRNA-mRNA targets with Olink protein pathways.
- Systems Biology Insights: Identification of convergent pathways where miRNA and protein biomarkers enhance predictive power.
- Customizable Panel Selection: Strategic guidance in selecting Olink panels (e.g., Inflammation, Cardiology) based on miRNA-targeted pathways.
End-to-End Translational Support
- Sample-to-Insight Services: Expertise in handling diverse biospecimens (tissue, plasma, serum) and managing cohort studies.
- Target Prioritization: Ability to cross-validate candidates using genetics and functional assays.
Technological Expertise and Platform Certification
- Advanced miRNA Detection: Expertise in low-abundance miRNA quantification with high sensitivity, especially for challenging biomarkers.
- Complementary Assay Integration: Ability to cross-validate results using orthogonal methods to eliminate technical biases.
- Olink CSP Certification: Proven capability to execute Olink workflows with high precision, and reproducibility, ensuring standardized data quality.
Sample Handling and Workflow Optimization
- Ultra-Low Sample Requirements: Support for minimal sample volumes (e.g., 1–8 μL for Olink; low plasma input for miRNA) critical for rare/clinical samples.
- Multi-Sample Compatibility: Expertise in diverse matrices (plasma, serum, exosomes, tissue lysates) and adaptability to sample-specific challenges.
Demo Results
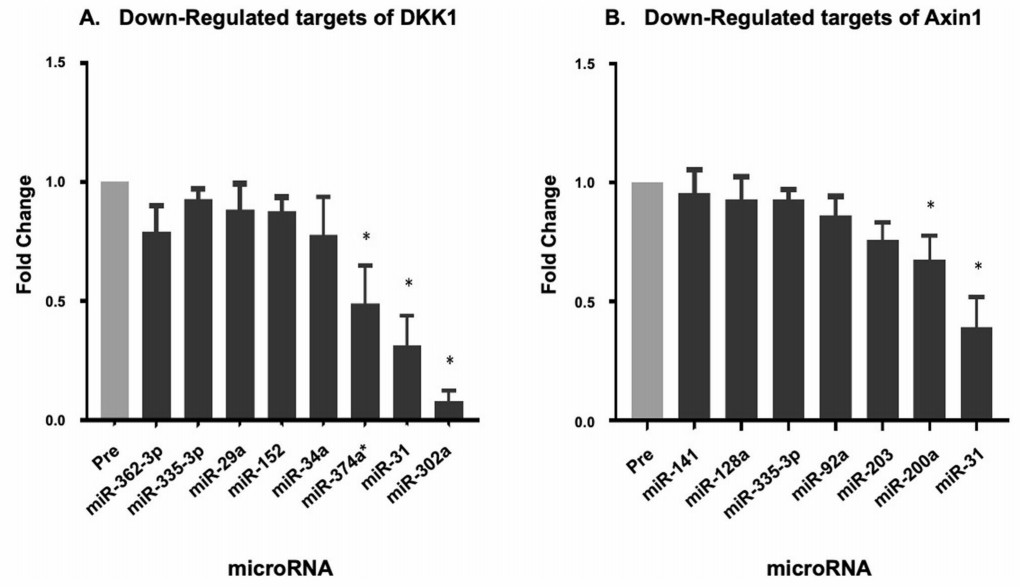 Figure 2: Downregulated miRNA which potentially target DKK1 and Axin1. (Twomey, L., et al. 2021)
Figure 2: Downregulated miRNA which potentially target DKK1 and Axin1. (Twomey, L., et al. 2021)
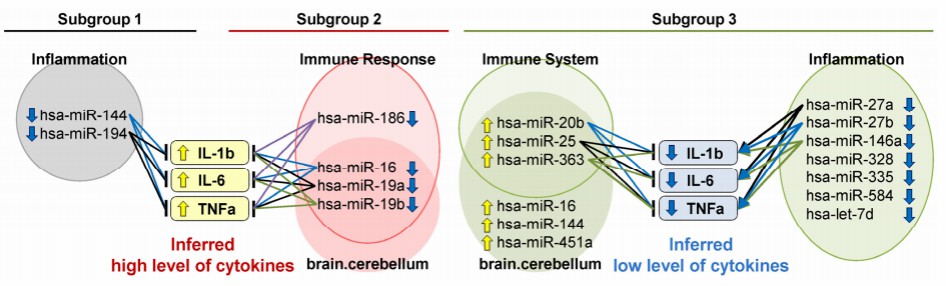 Figure 3: miRNA dysregulation drives inflammatory heterogeneity in schizophrenia, with Subgroup 2 representing a high-inflammation phenotype. (Miyano, T., et al. 2024)
Figure 3: miRNA dysregulation drives inflammatory heterogeneity in schizophrenia, with Subgroup 2 representing a high-inflammation phenotype. (Miyano, T., et al. 2024)
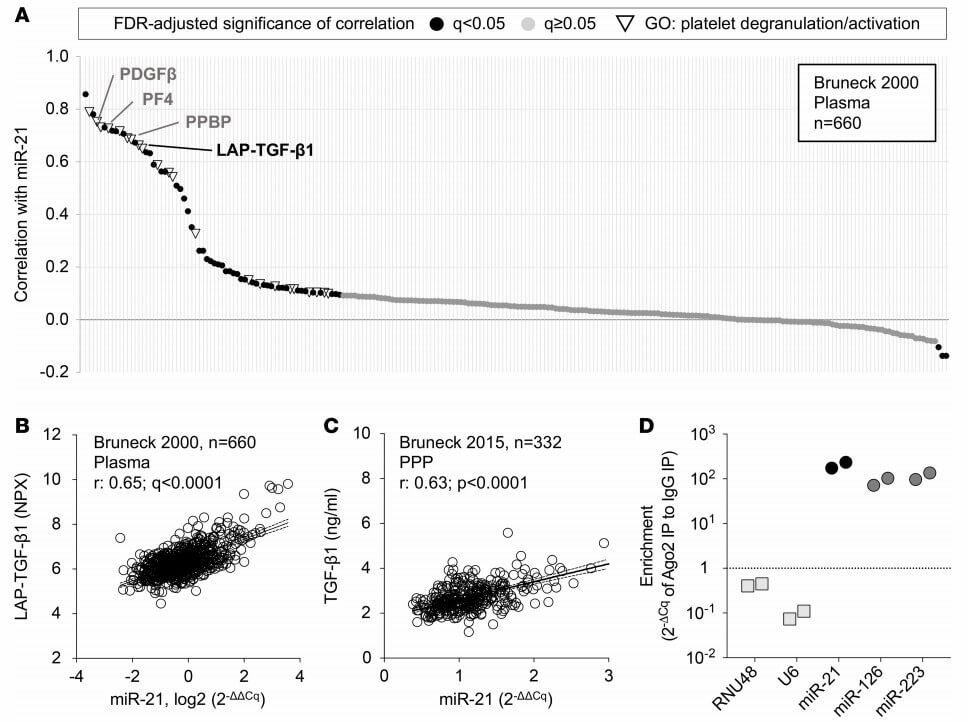 Figure 4: Plasma levels of miR-21 and TGF-β1. (Barwari, T., et al. 2018)
Figure 4: Plasma levels of miR-21 and TGF-β1. (Barwari, T., et al. 2018)
Sample Requirements
| Step | Volume/Amount | Preprocessing Requirements | ||
| Sample Types | Plasma | Olink: 1–5 µL miRNA: >500 µL |
Anticoagulant: EDTA/K₂EDTA (preferred), citrate (avoid heparin) | |
| Serum | Olink: 1–8 µL miRNA: >500 µL |
Clotting: 30–60 min at RT | ||
| CSF/Synovial Fluid | Olink: 4 µL miRNA: 200 µL |
Avoid repeated freeze-thaw | ||
| Tissue | Olink: 10–50 mg miRNA: 10–30 mg |
|
||
| Cell | Olink: ≥1×10⁶ cells miRNA: ≥5×10⁵ cells |
Lysis: RIPA buffer (Olink) / QIAzol (miRNA) | ||
| Exosomes | Olink: 100 µg protein miRNA: 50–100 µg |
|
||
| Step | Conditions | Maximum Duration | ||
| Storage | Pre-centrifugation: Blood: 4°C | ≤4 hours | ||
| Post-centrifugation: Plasma/Serum: −80°C | 2 years | |||
| Extracted RNA: −80°C (in RNase-free water) | 1 years | |||
| Tissue Homogenates: −80°C | 6 months | |||
| Transport | Dry ice (RNA/protein) or liquid N₂(tissue), ≤72 hours | |||
Notes
- Split samples into small volumes (e.g., 20 µL/tube) to avoid freeze-thaw cycles.
- Process all samples in a study using the same extraction kit/reagent lot.
FAQs
How are miRNA targets linked to Olink proteins?
A multi-step bioinformatic pipeline bridges miRNA targets and Olink-detected proteins. First, predicting miRNA-mRNA interactions uses databases to identify gene targets, followed by cross-referencing these genes with the proteins quantified via Olink' s panels. This is then supplemented by enrichment analysis to highlight pathways co-regulated by miRNA-protein pairs and validated using mechanistic studies to confirm direct repression of proteins.
Which sample types are compatible?
Olink and miRNA technologies support diverse biological samples. Olink assays is compatible with plasma, serum, cell supernatants, tissue lysates, cerebrospinal fluid, and exosomes. While miRNA analysis is optimized for serum/plasma, exosomes, fresh/frozen tissues, and cell lysates. Crucially, matched samples, such as aliquots from the same plasma or tissue extract, are strongly recommended for integrated studies. It enables direct correlation of miRNA expression with downstream protein biomarkers, minimizing technical variability. Special handling guidelines apply, including using EDTA/citrate anticoagulants for plasma-based Olink assays and avoiding hemolyzed samples for miRNA analysis to ensure data fidelity.
How is data integrated analytically?
Integrated analysis combines miRNA and Olink datasets through three key approaches:
- Correlation analysis identifies statistically significant inverse relationships.
- Pathway-centric mapping jointly overlays dysregulated miRNAs and proteins onto biological pathways, revealing cohesive regulatory networks.
- Multi-omics modeling incorporates RNA-seq or proteomics data where available, such as correlating Olink protein levels with corresponding mRNA expression to dissect post-transcriptional regulation.
What are common pitfalls to avoid?
Critical pitfalls include poor sample quality, technical batch effects, unaddressed confounders, and inadequate statistical power for multi-omics integration. Recommendations include pre-analytical validation of sample integrity, using unique molecular indices in miRNA sequencing, adjusting for biological variables in models, and validating miRNA-protein axes functionally to distinguish causal links from correlations.
References
- Twomey L, Navasiolava N, Robin A et al (2021) A dry immersion model of microgravity modulates platelet phenotype, miRNA signature, and circulating plasma protein biomarker profile. Sci Rep 11(1):21906.
- Miyano T, Mikkaichi T, Nakamura K et al (2024) Circulating microrna profiles identify a patient subgroup with high inflammation and severe symptoms in schizophrenia experiencing acute psychosis. International Journal of Molecular Sciences 25(8):4291.
- Barwari T, Eminaga S, Mayr U et al (2018) Inhibition of profibrotic microrna-21 affects platelets and their releasate. JCI Insight 3(21):e123335.