- Panel Features
- Panels List
- Workflow
- Why Creative Proteomics
- Demo
- Sample Requirements
- Case
- FAQ
- Sample Submission Pack
Customized and Ready-to-Use Olink Proteomics Panel for Body Fluid Sample
Why Analyze Body Fluid Samples?
1. Non-Invasive Accessibility –Body fluid enables straightforward, large-scale sample collection with minimal procedural complexity.
2. Systemic Biological Snapshot – Reflect real-time physiological states and dynamic molecular changes across multiple biological pathways.
3. Comprehensive Protein Capture – Detect circulating proteins, signaling molecules, and low-abundance biomarkers often undetectable in localized tissue analyses.
By leveraging body fluid proteomics, researchers obtain a holistic view of systemic biology, accelerating the discovery of mechanistically informative biomarkers for fundamental research.
What We Provide
Leveraging Olink's PEA technology, our platform enables high-sensitivity, high-throughput protein quantification
- Broad matrix coverage — Plasma, serum, CSF, saliva, urine, and cell culture supernatants with matrix-specific SOPs and stability guidance.
- Cohort & study design — Cross-sectional/longitudinal frameworks, randomization, batch-bridging calibrators, and kit/labeling & chain-of-custody support.
- Custom panels & signatures — 21–384+ plex target selection for inflammation, immune signaling, metabolism, neurology/oncology pathways; composite ratios and pathway indices.
- Low-input, high-throughput quantification — Minimal sample workflows with internal controls and replicate strategies tuned to each fluid.
- Olink + multi-omics integration — Seamless correlation with bulk/single-cell/spatial RNA-seq and metabolomics; pathway/network modeling and subgroup stratification.
- Biostatistics & reporting — NPX matrix with full QC; differential/time-course modeling, enrichment & co-expression; publication-ready figures and optional interactive dashboards.
- Orthogonal verification — ELISA/MSD, targeted LC-MS, and cross-matrix concordance checks to strengthen research conclusions.
This service is optimized for:
- Biomarker discovery and validation
- Comprehensive immune and inflammatory profiling
- Mechanistic investigations in oncology, cardiometabolism, and neurology research
- Scalable, ready-to-use workflows designed to accelerate proteomic research outcomes.
Features of the panel
- Species: Designed for human proteome profiling.
- Design: Target selection is facilitated through a user-friendly bioinformatics interface.
- Proteins: Supports 21- to 384+-plex panels targeting cytokines, immune responses, and specific pathways.
- Sample: 1 µL body fluid per analysis.
- Readout: Quantification via NPX (Normalized Protein Expression).
- Platform: Utilizes proprietary PEA technology on the Signature Q100 system.
- Exceptional Sensitivity: Detects low-abundance proteins down to pg/mL concentrations with high precision.
- Robust Reproducibility: Delivers consistent results with inter-assay coefficients of variation (CV) below 10%.
- Flexible Panel Options: Offers both pre-validated panels and fully customizable target selection for pathway-focused or broad-scale studies.
List of Our Body Fluid Proteome Assay Panel
Protein category
The Body Fluid Proteome Assay Panel quantitatively analyzes proteins spanning six core functional categories critical for biological investigation: Metabolic Regulators, Signal Transduction Proteins, Immune Response Modulators, Extracellular Matrix Proteins, Hormones and Carrier Proteins, and Cellular Stress Effectors (see Table. List of Body Fluid Biomarker Assay).
This panel encompasses fundamental biological processes, including intercellular communication, metabolic homeostasis, inflammatory regulation, and tissue reorganization, featuring precisely selected targets implicated in essential physiological pathways. Through systematic literature curation, we integrate rigorously validated biomarkers associated with nutrient sensing, cellular adaptive responses, and systemic physiological balance, delivering a comprehensive platform for proteome-scale exploration across research domains such as metabolic physiology, aging mechanisms, and environmental adaptation studies. The panel's structured design facilitates focused examination of protein interaction networks while preserving the analytical depth necessary for mechanistic research.
Table. List of Body Fluid Biomarker Assays
Protein Functions
Biological process
primarily involved in key biological processes including metabolic pathway regulation, intracellular and intercellular signaling cascades, cellular stress adaptation mechanisms, immune response coordination, and tissue microenvironment homeostasis.
Disease area
primarily utilized in research areas involving metabolic dysregulation studies, chronic inflammatory model systems, age-related biological processes, cellular transformation mechanisms, and neurological adaptation models.
The Application of Body Fluid Proteome Assay.
The Body Fluid Proteome Assay enables comprehensive quantification of protein biomarkers across diverse biological pathways, providing researchers with a versatile tool for:
- Systematic profiling of immune and inflammatory responses in experimental model systems;
- Mechanistic exploration of metabolic adaptation and nutrient signaling pathways in physiological studies.
- Identification of extracellular matrix and tissue remodeling markers in fibrosis and wound healing research.
- Discovery of protein signatures associated with cellular stress and adaptive responses in environmental exposure models.
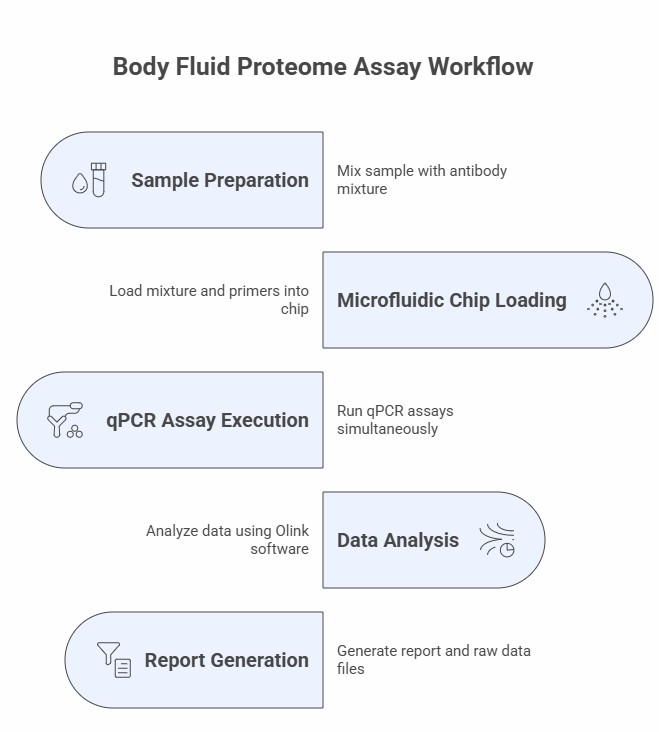
Workflow of Olink Proteomics
Why Creativ Proteomics
Pathway-Focused Bioinformatics
Preconfigured workflows for metabolic, inflammatory, and stress-response pathways, including 50+ validated biomarker ratios for mechanistic insights.
Broad Sample Compatibility
Validated for plasma, serum, CSF, saliva, and cell culture supernatants, with optimized protocols for each matrix.
Multi-Omics Integration
Standardized data outputs compatible with transcriptomic and metabolomic platforms, enabling unified pathway mapping and co-expression analysis.
Transparent Quality Metrics
Comprehensive documentation of pre-analytical variables, LOD/LOQ, and stability data for robust experimental planning.
Demo Results of Olink Data
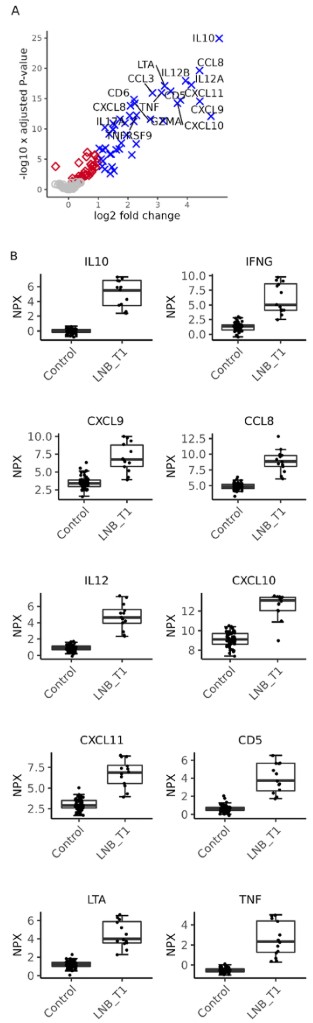 Figure 1: Differentially expressed proteins in patients diagnosed with Lyme neuroborreliosis (LNB) compared to the control group. (Haglund, S., et al. 2025)
Figure 1: Differentially expressed proteins in patients diagnosed with Lyme neuroborreliosis (LNB) compared to the control group. (Haglund, S., et al. 2025)
Sample Requirements
| Sample Type | Recommended Sample Size | Sample Quality | Pre-treatment and Storage | Sample Transport |
| Plasma/Serum/Body Fluid | The minimum input volume per assay is 40 µL. | For optimal results, maintain concentrations between 0.5 and 1 mg/mL. | For long-term preservation, aliquot samples and maintain at -80°C. | Ship sealed samples on dry ice (-80°C). |
| Tissue | ||||
| Cells | ||||
| Exosomes | ||||
| Other |
Case Study
Cerebrospinal Fluid Biomarkers in Opioid Dependence: Evidence of Neuroimmune Activation and Ion Composition Changes, Without Alteration in Orexin-A
Journal: Addiction biology,
Year: 2025
- Background
- Methods
- Results
Opioid abuse is a serious global health challenge, leading to increased morbidity, mortality, and societal costs. This study aims to explore neuroinflammation, neuronal damage, and potential changes in the orexin system or β-amyloid metabolism in the cerebrospinal fluid (CSF) of individuals undergoing opioid replacement therapy (OST). This cross-sectional study compared cerebrospinal fluid biomarkers in individuals undergoing OST with controls.
Cerebrospinal fluid samples were collected using a 25-G non-injured needle by lumbar puncture performed in an upright sitting position. Collection takes place during the day and ranges from 9 a.m. to 1 p.m. The collected CSF samples were centrifuged at a rotational rate of 2000g for 10 minutes at room temperature to remove cells and debris, and then aliquoted in a container at -80°C for subsequent biochemical analysis. Tyrosine protein kinase receptor 3 (TYRO3), tyrosine kinase receptor Axl (AXL), tyrosine kinase Mer (MER), and growth arrest-specific protein 6 (GAS6) were analyzed using Olink Proteomics' Olink Explore 3072 platform.
Each participant provided a thorough medical history, including medication use within the past 6 months, followed by a lumbar puncture to obtain a CSF sample. These samples contain molecules associated with neuroinflammation, neuronal and glial damage, and β-amyloid. Metabolic and orifice peptide function in the participants' cerebrospinal fluid was analyzed, while electrolyte levels were also detected. Specifically, we analyzed sTREM-2, YKL-40, IL-1β, IL-6, IL-8, IL-10, TNF-α, AXL, MER, TYRO3, GAS6, NfL, GFAP, total tau (T-tau), phosphorylated tau (P-tau), neurogranulin protein, Aβ40, Aβ42, Aβ42/Aβ40 ratio, orifice peptide A, sPDGFR-β, and electrolytes. The study included 15 control subjects and 17 subjects in the opioid replacement treatment group. Patients receiving opioid replacement therapy have elevated levels of sTREM-2, Aβ42/Aβ40, and NfL in the cerebrospinal fluid. Conversely, the concentrations of sodium and chloride ions were lower compared to the control group. No significant differences were found between groups in other biomarkers, including Aureixin-A. However, when standardized to Aβ40 levels, YKL-40, IL-8, TYRO3, and P-τ were also elevated in individuals with opioid dependence. Elevated markers of neuroimmune activation, neuronal damage, and β-amyloid metabolism in opioid dependence suggest that CNS inflammation is a factor in its pathological mechanism. A decrease in electrolyte levels means a disturbance in the regulation of water in the cerebrospinal fluid, which may be associated with impaired glial function. These findings highlight neural and non-neural mechanisms in opioid dependence.
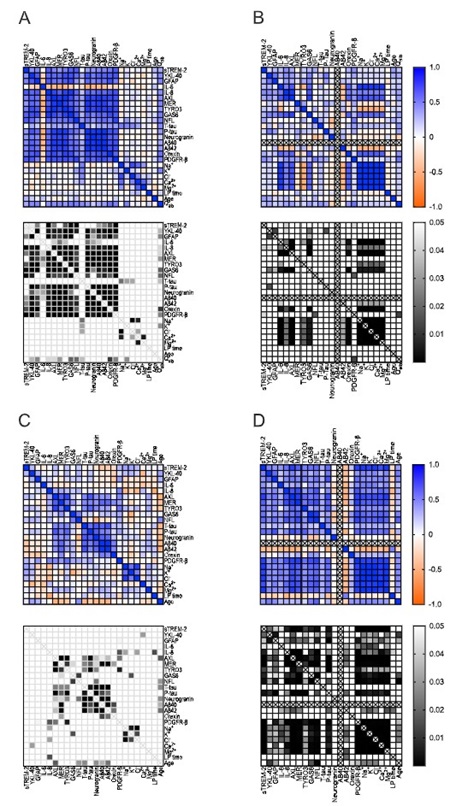 Figure 1. Correlation between cerebrospinal fluid biomarkers between opioid-dependent and control groups (with or without standardization of Aβ40). ( Lyckenvik, T, et al. 2025)
Figure 1. Correlation between cerebrospinal fluid biomarkers between opioid-dependent and control groups (with or without standardization of Aβ40). ( Lyckenvik, T, et al. 2025)
FAQs
What types of biological fluids are compatible with Olink's biomarker panels for research applications?
Olink's assays support a wide range of body fluids, including plasma, serum, cerebrospinal fluid (CSF), saliva, and cell culture supernatants. Each fluid type undergoes rigorous validation to ensure data reproducibility, with detailed protocols provided for sample collection, storage, and preprocessing to minimize pre-analytical variability.
How does Olink ensure high specificity and sensitivity in protein detection for low-abundance biomarkers?
The platform utilizes Proximity Extension Assay (PEA) technology, which combines dual antibody recognition with DNA-based amplification, enabling ultrasensitive detection (sub-pg/mL range) even in complex matrices. This minimizes cross-reactivity and enhances specificity for quantifying low-abundance proteins in research samples.
Can researchers customize biomarker panels for specific experimental pathways or disease mechanisms?
Yes, Olink offers tailored panel configurations—from focused 20-plex panels to broad 500+ protein profiles—allowing researchers to select targets relevant to their study (e.g., inflammation, oncology, or neurology pathways). Custom panels can include proprietary targets or synthetic controls upon request.
What bioinformatics support is available for proteomic data analysis and integration with other omics datasets?
Olink provides dedicated software tools for quality control, differential expression analysis, and pathway enrichment. Data outputs are compatible with major transcriptomic/metabolomic platforms, enabling multi-omics correlation studies through co-expression networks and multivariate visualization.
References
- Haglund, S., Gyllemark, P., Forsberg, P., et al. (2025). Cerebrospinal fluid protein profiling of inflammatory and neurobiological markers in Lyme neuroborreliosis. Scientific reports, 15(1), 20190. https://doi.org/10.1038/s41598-025-06146-y
- Lyckenvik, T., Woock, M., Johansson, K., et al. (2025). Cerebrospinal Fluid Biomarkers in Opioid Dependence: Evidence of Neuroimmune Activation and Ion Composition Changes, Without Alteration in Orexin-A. Addiction biology, 30(6), e70053. https://doi.org/10.1111/adb.70053