- Panel Features
- Panels List
- Workflow
- Why Creative Proteomics
- Demo
- Sample Requirements
- Case
- FAQ
- Sample Submission Pack
Customized and Ready-to-Use Olink Proteomics Panel for Cell Culture Sample
Why Analyze Cell Culture Samples?
1. Controlled Experimental Systems – Cell cultures provide defined, reproducible environments ideal for manipulating variables and studying specific biological mechanisms without systemic interference.
2. Direct Cellular Response Monitoring – Enables real-time investigation of protein expression changes, signaling pathway activation, and secretory profiles in response to experimental stimuli.
3. High-Resolution Molecular Access – Facilitates detection of intracellular proteins, transient phosphorylation events, and low-abundance targets often obscured in complex biofluids or tissues.
By leveraging cell culture proteomics, researchers achieve precise dissection of cellular mechanisms, accelerating the discovery of functionally relevant biomarkers for fundamental biological research.
What We Provide
- Model-focused programs — Primary/immortalized cells, iPSC-derived cells, organoids; mono-, co-, and tri-culture setups under genetic or chemical perturbations.
- Sample coverage — Conditioned media (secretome) and cell lysates with serum-free/low-serum–compatible SOPs; supports time-course and dose-response series.
- Custom panels & study design — 21–384+ plex selection for cytokines/chemokines, growth factors, signaling, stress/UPR, apoptosis, ECM/EMT; plate randomization and batch-bridging controls.
- Low-input, high-throughput — 10–20 μL per well; 96/384-well workflows with media blanks, spike-in recovery checks, and replicate strategy.
- Olink + multi-omics integration — Optional correlation with bulk/single-cell/spatial RNA-seq, imaging, and metabolomics for pathway/network insights.
- Biostatistics & reporting — NPX matrix + full QC; differential/time-series analyses, enrichment and co-expression networks; publication-ready figures and optional interactive dashboards.
- Orthogonal verification — ELISA/MSD, targeted LC-MS, and Western blot to technically confirm key findings.
This service is optimized for:
- Secreted biomarker discovery and validation
- Cell signaling and pathway activation studies
- In vitro model validation and mechanistic screening
- High-throughput, ready-to-use workflows for cell culture proteomics
Features of the panel
- Species: Human Proteome Optimized.
- Design: Intuitive bioinformatics interface for target selection.
- Proteins: Supports 21- to 384+-plex panels targeting cytokines, immune responses, and specific pathways.
- Sample: 1-6 µL cell culture per analysis.
- Readout: Quantification via NPX (Normalized Protein Expression).
- Platform: Utilizes proprietary PEA technology on the Signature Q100 system.
- High Sensitivity: Detects low-abundance secreted proteins and signaling molecules in the pg/mL range.
- Excellent Reproducibility: Maintains inter-assay coefficients of variation (CV) below 10% for consistent results.
- Flexible Panel Options: Offers pre-validated panels or fully customizable target selection tailored to cell signaling, secretion, or response studies.
List of Our Cell Culture Proteome Assay Panel
Protein category
The Cell Culture Proteome Assay Panel quantitatively analyzes proteins across critical functional categories for in vitro research, including Secreted Signaling Proteins, Surface Receptors, Metabolic Enzymes, Apoptosis Regulators, Growth Factors, and Stress Response Mediators (see Table: List of Cell Culture Biomarker Assay).
This panel encompasses fundamental cellular processes such as intercellular communication, nutrient metabolism, proliferation signaling, and adaptive responses to environmental stimuli, featuring strategically selected targets implicated in key mechanistic pathways. Through rigorous literature curation and experimental validation, we incorporate scientifically relevant biomarkers associated with cell-state transitions, secretory profiles, and pathway activation states, providing researchers with a specialized tool for comprehensive proteome investigations in diverse in vitro research areas, including drug response studies, cellular differentiation models, and microenvironment interaction mechanisms. The panel's optimized design enables targeted analysis of condition-specific protein networks while maintaining the quantitative precision required for mechanistic discovery in cell-based systems.
Table. List of Cell Culture Biomarker Assays
Protein Functions
Biological process
The Cell Culture Proteome Assay provides a powerful platform for investigating fundamental biological processes in in vitro systems, enabling detailed quantification of proteins central to metabolic pathway regulation, intracellular signal transduction, and intercellular communication networks.
Disease area
The Cell Culture Proteome Assay serves as a foundational research tool for modeling disease mechanisms in vitro, enabling the investigation of pathological processes such as dysregulated metabolic flux in mitochondrial disorders, aberrant signaling pathways in neurodegenerative models, maladaptive stress responses in environmental toxin exposure, disrupted immune coordination in autoimmune-like conditions, and loss of cellular homeostasis in fibrotic or inflammatory microenvironments.
The Application of Cell Culture Proteome Assay.
The Cell Culture Proteome Assay enables high-resolution quantification of protein dynamics in in vitro systems, offering researchers a powerful tool for:
- Mechanistic investigation of cell signaling pathways and receptor activation under controlled experimental conditions;
- Discovery of protein biomarkers associated with cellular stress responses (e.g., hypoxia, nutrient deprivation, oxidative stress) in environmental or pharmacological studies;
- Real-time monitoring of immune cell crosstalk and cytokine network interactions in co-culture systems;
- Integration with multi-omics datasets (e.g., transcriptomics, metabolomics) for holistic functional biology insights in in vitro research.
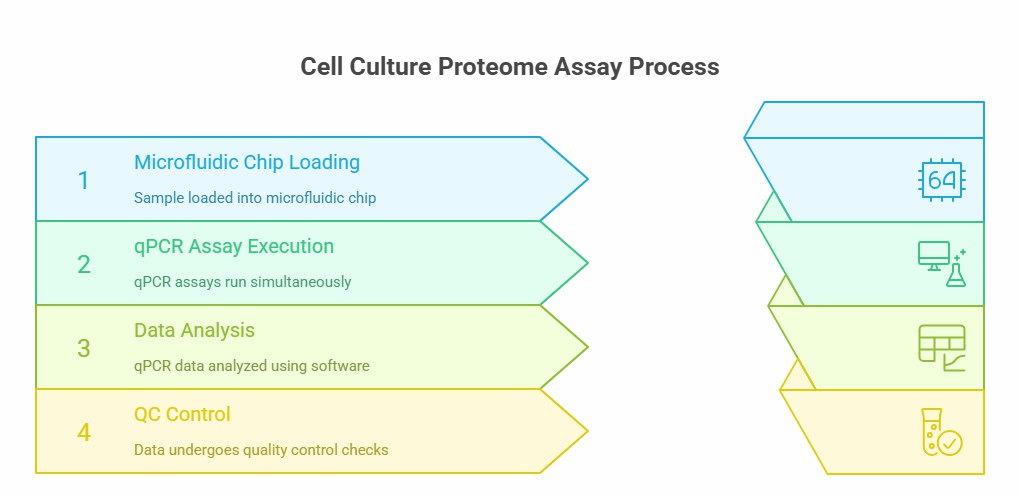
Workflow of Olink Proteomics
Why Creativ Proteomics
Specialized Secretome & Lysate Analysis
Panels are specifically optimized for the unique matrices of cell culture supernatants and lysates, providing robust quantification of secreted factors and intracellular proteins.
Ultra-Sensitive Detection for Low-Abundance Targets
PEA technology achieves sub-pg/mL sensitivity, enabling reliable detection of dilute cytokines, growth factors, and signaling proteins in conditioned media without sample concentration.
Preconfigured In Vitro Pathway Workflows
Access dedicated bioinformatics workflows for common cell culture applications, including cytokine storm modeling, compound mechanism-of-action studies, and stress-response pathway activation.
Media-Compatibility Validation
Assays are validated for performance across diverse culture conditions, including serum-free, low-serum, and complex supplemented media, with minimal interference.
Demo Results of Olink Data
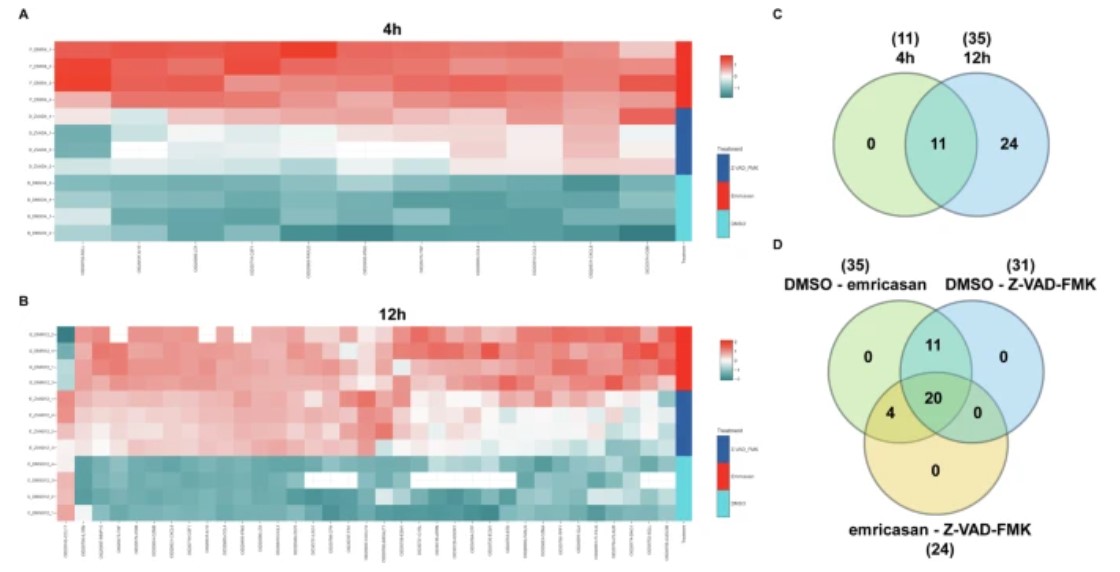 Figure 1: Secretome analysis of natural killer cells treated with pancaspase inhibitors. (Hajikhezri, Z., et al. 2025)
Figure 1: Secretome analysis of natural killer cells treated with pancaspase inhibitors. (Hajikhezri, Z., et al. 2025)
Sample Requirements
| Sample Type | Recommended Sample Size | Sample Quality | Pre-treatment and Storage | Sample Transport |
| Cells culture | The assay requires a 40 µL minimum sample volume. | Optimal performance is achieved at concentrations of 0.5–1 mg/mL. | For long-term stability, store aliquoted samples at -80°C. | Ship sealed specimens on dry ice (-80°C).> |
Case Study

Colorectal Cancer Cells–Derived Exosomal PIK3CA Mutation DNA Promotes Tumor Metastasis by Activating Fibroblast and Affecting Tumor Metastatic Microenvironment
Journal: Advanced science
Year:2025
- Background
- Methods
- Results
Exosomes are involved in the formation of the tumor metastasis microenvironment (TME) by transmitting tumor-specific substances. However, most of the current research has focused on the RNA and proteins of exosomes, while relatively little research has been done on the DNA of exosomes.
MEFs (mouse embryonic fibroblasts) were cultured in 6-well plates. Exosomes extracted from wild-type and mutant cells were administered continuously for 72 hours, and when the cells reached approximately 70% adherent density the next day. Subsequently, cell cultures from each group were collected after 72 hours of incubation in serum-free medium. These cell cultures were then submitted to Applied Protein Technology Biotechnology for Olink targeted proteomics analysis.
The main role of fibroblasts is to produce paracrine cytokines, which in turn promote tumor metastasis. The investigators hypothesize that specific factors released by fibroblasts contribute to colorectal cancer metastasis. To determine the levels of cytokines, they used Olink's proteome-specific assay to identify extracellular matrix (ECM) from MEFsWT and MEFsMT. Approximately 40 cytokines are secreted, 7 of which include colony-stimulating factor (CSF), recombinant macrophage-derived chemokine (MDC), IL6, macrophage inflammatory protein-1 (MIP-1), interferon, α2 (IFNα2), tumor necrosis factor (TNF), and stromal cell-derived factor-1 (SDF-1), exhibit significantly elevated protein levels in the extracellular matrix of MEFsMT (Figure 5A).
 Figure 1. Olink targets proteome testing as well as different types of bone marrow mesenchymal stem cells (CMs). (Wang, R., et al. 2025)
Figure 1. Olink targets proteome testing as well as different types of bone marrow mesenchymal stem cells (CMs). (Wang, R., et al. 2025)
FAQs
What types of cell culture applications is this platform designed for?
The technology is ideal for in vitro research applications, including mechanistic studies of cell signaling pathways, comprehensive profiling of secretome changes in response to compound treatment, analysis of immune cell crosstalk in co-culture systems, and biomarker discovery for cellular adaptations to stress or genetic manipulation.
What sample types from cell culture are compatible with the assay?
The platform is specifically validated for cell culture supernatants and lysates. It demonstrates consistent performance across various media formulations (including serum-free and reduced-serum conditions) and provides optimized protocols for sample preparation to ensure compatibility and maximize data quality.
How does the platform address the challenge of low-abundance biomarkers in cell culture media?
Utilizing Proximity Extension Assay (PEA) technology, the platform achieves sub-pg/mL sensitivity, enabling robust detection of low-abundance secreted factors and signaling proteins even in the complex background of cell culture media, without the need for pre-concentration steps that can introduce variability.
Can we design a custom panel for a specific cellular pathway or experimental model?
Yes. Researchers can configure tailored panels from a validated library of over 3,500 human protein targets. This allows for the creation of focused panels targeting specific pathways (e.g., cytokine signaling, apoptosis, angiogenesis) relevant to you in vitro model system, while maintaining the platform's high reproducibility standards.
References
- Hajikhezri, Z., Zygouras, I., Sönnerborg, A., et al. (2025). Pan-caspase inhibitors induce secretion of HIV-1 latency reversal agent lymphotoxin-alpha from cytokine-primed NK cells. Cell death discovery, 11(1), 44. https://doi.org/10.1038/s41420-025-02330-1
- Wang, R., Li, W., Lv, Y., et al. (2025). Colorectal Cancer Cells-Derived Exosomal PIK3CA Mutation DNA Promotes Tumor Metastasis by Activating Fibroblast and Affecting Tumor Metastatic Microenvironment. Advanced science (Weinheim, Baden-Wurttemberg, Germany), 12(27), e2501792. https://doi.org/10.1002/advs.202501792