- Panel Features
- Panels List
- Workflow
- Why Creative Proteomics
- Demo
- Sample Requirements
- Case
- FAQ
- Sample Submission Pack
Customized and Ready-to-Use Olink Proteomics Panel for Oncology
Why Analyze Oncology?
1. Non-Invasive Biomarker Access – Plasma and serum provide easily obtainable samples for large-scale cancer studies, enabling longitudinal monitoring of tumor dynamics without invasive procedures.
2. Systemic Tumor Insights – Reflect real-time changes in tumor-microenvironment interactions, immune responses, and metabolic adaptations relevant to cancer progression.
3. Dynamic Protein Profiles – Capture circulating tumor-derived proteins, signaling molecules, and immune markers that offer complementary data to tissue-based analyses.
By leveraging oncology-focused proteomics, researchers gain critical insights into tumor biology and host-tumor interactions, accelerating the discovery of mechanistically informative biomarkers for cancer biology studies.
What We Provide
- Cancer biology–focused programs — Solid tumors and hematologic malignancies; projects centered on tumor signaling, tumor microenvironment (TME), metastasis biology, DNA-damage response, angiogenesis, and cellular stress.
- Samples & preclinical models — Plasma/serum and other biofluids, optional matched FFPE/fresh-frozen tissue and PBMCs; support for cell lines, organoids, mouse and PDX studies; longitudinal research cohorts.
- Custom panels & study design — 21–384+ plex target selection (checkpoints, cytokines, ECM/EMT, DDR, metabolism); hypothesis matrices, randomization, batch-bridging controls; pilot feasibility runs.
- Olink + multi-omics integration — Bulk/single-cell/spatial RNA-seq, WES/WGS, proteogenomic mapping; pathway/network analysis, unsupervised clustering, subtype discovery.
- Biostatistics & reporting — NPX matrix with full QC; differential and longitudinal modeling, enrichment analyses, co-expression networks; publication-ready figures and optional interactive dashboards.
- Orthogonal verification — ELISA/MSD, targeted LC-MS, IHC/qPCR to technically confirm candidates and strengthen research conclusions.
This service is optimized for:
- Discovery and validation of oncology biomarkers
- Comprehensive profiling of tumor-immune interactions and signaling pathways
- Mechanistic investigations of cancer progression and treatment response mechanisms
- Scalable, ready-to-use workflows for cancer proteomics research
Features of the panel
- Species: Optimized for human proteome profiling in cancer studies.
- Design: Customizable target selection via bioinformatics interface for tumor pathways.
- Proteins: 21- to 384-plex panels targeting immune checkpoint markers, tumor antigens, and microenvironment factors
- Sample: 1-6 µL plasma/serum per analysis.
- Readout: NPX-based quantification for normalized protein expression.
- Platform: PEA technology ensuring sensitive detection
- Exceptional Sensitivity: Detects low-abundance oncology markers (e.g., immune checkpoint proteins, tumor antigens) down to pg'ml, concentrations with high precision.
- Flexible Panel Options: Offers pre-validated panels targeting cancer pathways (e.g., tumor microenvironment immune evasion) and fully customizable target selection for specific research needs.
List of Our Oncology Proteome Assay Panel
Protein category
The Oncology Proteome Assay Panel quantitatively analyzes proteins across six critical functional categories for cancer research: Tumor Signaling Proteins, Immune Checkpoint Regulators, Tumor Microenvironment Components, Metastasis-Related Factors, DNA Damage Response Proteins, and Angiogenesis Regulators (see Table: List of Oncology Biomarker Assays).
This panel encompasses fundamental biological processes, including cell proliferation control, immune evasion mechanisms, extracellular matrix remodeling, hypoxic stress responses, and cellular invasion pathways, featuring strategically selected targets implicated in oncogenic transformation and cancer progression mechanisms. Through systematic literature curation and experimental validation, we incorporate rigorously vetted biomarkers associated with tumor suppressor pathways, oncogenic signaling, immune cell infiltration, and treatment resistance mechanisms, providing researchers with a specialized tool for comprehensive proteome investigations in diverse cancer research areas, including tumor biology studies, immunotherapy development, and metastatic process research. The panel's optimized design enables focused examination of cancer-related protein interaction networks while preserving the analytical depth required for mechanistic discovery in oncology research.
Table. List of Oncology Biomarker Assays
Protein Functions
Biological process
primarily involved in key biological processes including cell cycle regulation, apoptosis control, angiogenesis modulation, immune response regulation, and cellular adhesion mechanisms, enabling comprehensive investigation of tumor development and progression pathways in cancer research models.
Disease area
primarily utilized in research areas involving tumor microenvironment studies, metastasis mechanisms, drug resistance pathways, cancer immunotherapy development, and tumor-stroma interactions, providing investigators with essential tools for exploring fundamental biological mechanisms underlying cancer biology and treatment responses.
The Application of Oncology Proteome Assay.
The Oncology Proteome Assay enables high-throughput quantification of protein biomarkers across cancer-related pathways, providing researchers with a powerful tool for:
- Discovery of protein networks underlying tumor immune evasion and metastatic mechanisms in cancer models;
- Mechanistic investigation of therapy resistance pathways and DNA damage response in treatment studies;
- Identification of molecular signatures associated with tumor microenvironment remodeling and angiogenesis;
- Analysis of cellular proliferation signaling and apoptosis regulation in experimental oncology research.
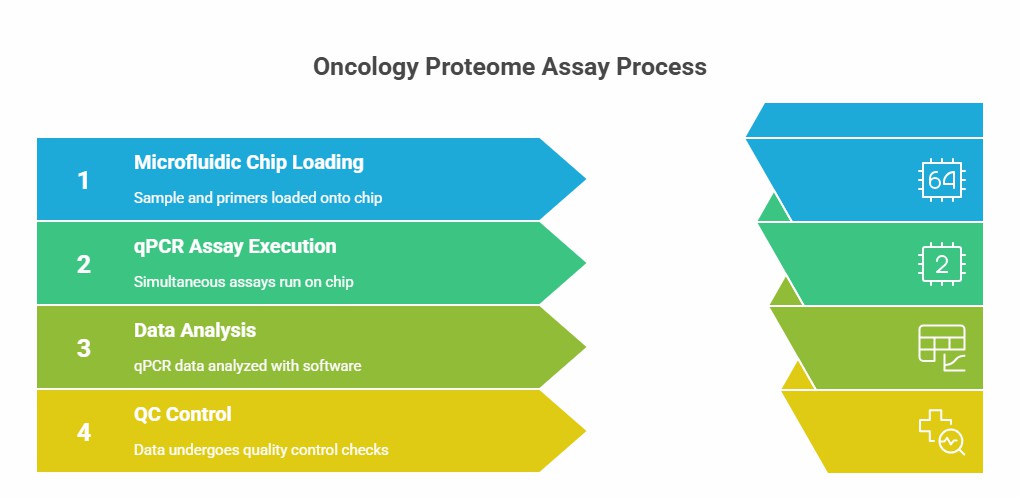
Workflow of Olink Proteomics
Why Creativ Proteomics
Rigorous Quality Control
Implements 12-stage QC protocols with batch-effect correction and sample integrity validation, ensuring <8% CV for 90% of oncology-specific biomarkers.
Oncology-Specific Analytical Frameworks
Preconfigured workflows for tumor-immune interactions, metastasis mechanisms, and therapy resistance studies with validated biomarkers.
High Sensitivity for Tumor Targets
Detects low-abundance biomarkers (e.g., immune checkpoint proteins, tumor antigens) at concentrations as low as 0.2 pg/mL with <15% CV.
Pathway-Centric Bioinformatics
Automated tools for cancer pathway enrichment analysis (e.g., angiogenesis, apoptosis, DNA repair).
Demo Results of Olink Data
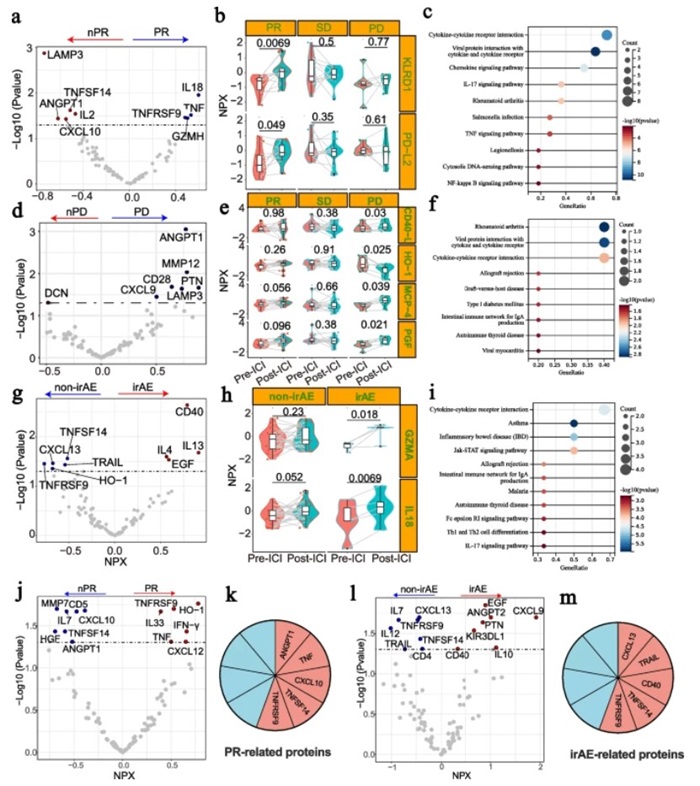 Figure 1: Plasma proteomics studies based on immune checkpoint inhibitor response and toxicity. (Gao, Y., et al. 2025)
Figure 1: Plasma proteomics studies based on immune checkpoint inhibitor response and toxicity. (Gao, Y., et al. 2025)
Sample Requirements
| Sample Type | Recommended Sample Size | Sample Quality | Pre-treatment and Storage | Sample Transport |
| Plasma/Serum/Body Fluid | The minimum input volume per assay is 40 µL. | For optimal results, maintain concentrations between 0.5 and 1 mg/mL. | For long-term preservation, aliquot samples and maintain at -80°C. | Ship sealed samples on dry ice (-80°C). |
| Tissue | ||||
| Cells | ||||
| Exosomes | ||||
| Other |
Case Study
Association of circulating immuno-oncology biomarkers with breast cancer risk: insights from two prospective cohorts
Journal: NPJ precision oncology
Year: 2025
- Background
- Methods
- Results
Immuno-oncology biomarkers are promising tools for assessing cancer risk and early detection. To determine and validate the association between these markers and breast cancer, the investigators conducted a nested case-control study in the Taizhou Longitudinal Study (TZL) cohort and the UK Biobank Pharmacomics Project (UKB-PPP), which included a total of 195 and 881 patients with new-onset breast cancer and their matched controls.
For TZL, a baseline plasma sample is taken and stored at -80°C for subsequent analysis. The analytical tool used is the Olink Target Immuno-Oncology panel, which contains 92 unique protein biomarkers. Before analysis, samples are thawed in a controlled gradient and randomly assigned to a 96-well plate containing 35 microliters of plasma per well. These prepared samples were then transported in dry ice to the Olink laboratory in Shanghai, China. Quantification of protein levels is achieved by proximity extension assay (PEA) technology, which generates standardized protein expression (NPX) values for each protein. To maintain the integrity of the experiment and ensure sample quality, four internal controls are incorporated into each sample. The quality control (QC) process includes evaluating the standard deviation of these internal controls on each sample plate. Only boards with a standard deviation of less than 0.2 NPX are considered eligible samples for reportable data. In addition, the quality of the samples was assessed by comparing the deviation values of each sample to the controls. If the deviation from the median value is less than 0.3 NPX, the sample is considered to have passed the quality control. After quality control, 33 samples (20 cases, 13 controls) were excluded. NPX values below the lower limit of detection (LOD) are retained. The final dataset contains 195 matching pairs for analysis.
After adjusting for potential confounders and setting an FDR (false positive rate) threshold of less than 0.05, 11 of the 92 immuno-oncology proteins were significantly associated with breast cancer. For 10 proteins exhibiting positive correlations, adjusted odds ratios (OR) ranged from 1.38 (C-X-C motif chemokine 11, CXCL11: 95% confidence interval = 1.11 - 1.72) to 3.71 (CASP8: 95% confidence interval = 2.49 - 5.54) for each increase in protein levels; Matrix metalloproteinase-7 (MMP7) showed a negative correlation (OR = 0.69, 95% confidence interval = 0.55 - 0.87) (Fig.1).
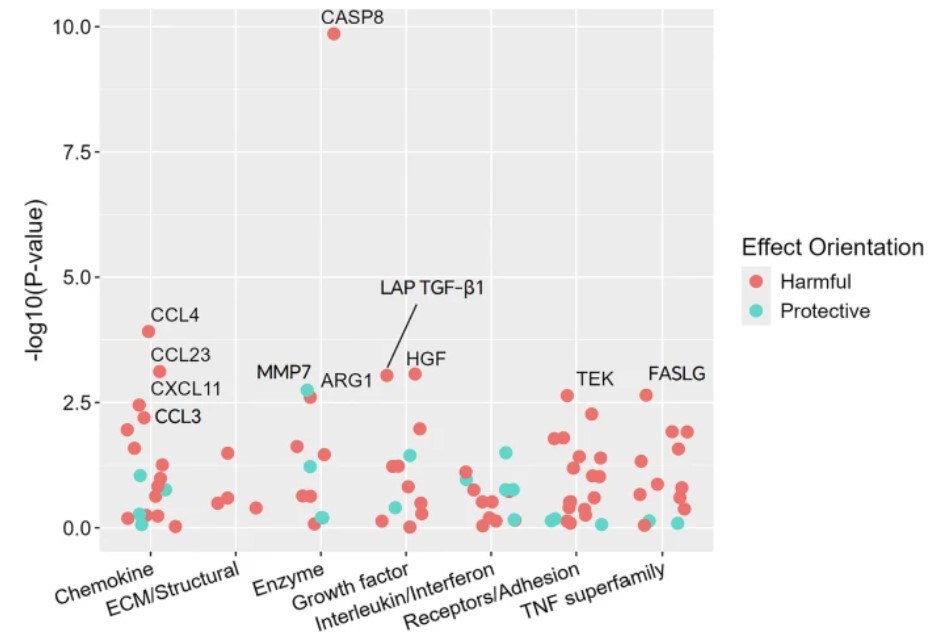 Figure 1. Targeted detection of the association between 92 proteins in the panel and breast cancer incidence. (Zheng, J. et al. 2025)
Figure 1. Targeted detection of the association between 92 proteins in the panel and breast cancer incidence. (Zheng, J. et al. 2025)
FAQs
What research applications does Oncology Proteome assay support?
The panels enable investigation of tumor-immune interactions, therapy resistance mechanisms, metastatic pathways, and tumor microenvironment remodeling in cancer models.
What sample types are compatible with oncology panels?
The assay supports plasma, serum, and cell culture supernatants from cancer models, with optimized protocols for each matrix to ensure reproducibility in oncology studies.
How does the technology achieve high specificity for cancer biomarkers?
The platform employs PEA technology with dual antibody recognition, enabling precise detection of low-abundance targets like immune checkpoint proteins and tumor-derived antigens
References
- Gao, Y., Qi, F., Zhou, W., et al. (2025). Liquid biopsy using plasma proteomics in predicting efficacy and tolerance of PD-1/PD-L1 blockades in NSCLC: a prospective exploratory study. Molecular biomedicine, 6(1), 51. https://doi.org/10.1186/s43556-025-00291-6
- Zheng, J., Suo, C., Wu, Y., et al. (2025). Association of circulating immuno-oncology biomarkers with breast cancer risk: insights from two prospective cohorts. NPJ precision oncology, 9(1), 238. https://doi.org/10.1038/s41698-025-01019-z