- Panel Features
- Panels List
- Workflow
- Why Creative Proteomics
- Demo
- Sample Requirements
- Case
- FAQ
- Sample Submission Pack
Customized and Ready-to-Use Olink Proteomics Panel for Metabolic Diseases
Why Focus on Metabolic Diseases Proteomics?
1. Systemic Metabolic Regulation– Capture circulating factors governing energy homeostasis, nutrient sensing, and hormonal signaling across tissues.
2. Dynamic Pathway Activity Monitoring– Track real-time fluctuations in inflammatory mediators, adipokines, and lipid metabolites reflecting metabolic status.
3. Multi-Organ Communication Analysis – Reveal inter-organ crosstalk through secreted proteins critical in metabolic synchrony and dysregulation.
By leveraging proteomic approaches in metabolic disease research, investigators gain actionable insights into network-level biological mechanisms driving conditions like insulin resistance, dyslipidemia, and ectopic lipid accumulation.
What We Provide
- Panel selection & setup — Recommend suitable Olink Target/Explore/Flex panels and finalize 21–384+ analytes for adipokines, hepatokines, myokines, insulin signaling, lipid metabolism, and low-grade inflammation.
- Assay execution with strict QC — PEA on plasma/serum (1–6 μL) using internal/plate controls, replicate checks, and detection/call-rate thresholds.
- NPX data processing — Inter-plate normalization/bridging, LOD handling with flags, and batch effect review.
- Analysis on your provided labels/metadata — Group comparisons, correlations with client-supplied metrics (e.g., HbA1c, HOMA-IR, TG/HDL), and pathway/module enrichment across metabolic axes.
- Reporting & handoff — Deliver NPX matrix (.csv), full QC dossier, and publication-ready figures (volcano/heatmap/pathway tables); reusable methods paragraph.
- Optional — Overlay with client-provided metabolomics/lipidomics/transcriptomics; orthogonal verification (ELISA/MSD, targeted LC-MS).
Optimized for Metabolic Research
- Insulin resistance mechanism studies
- Lipid metabolism dysregulation analysis
- Adipose tissue inflammation profiling
- Mitochondrial function assessment
Features of the panel
- Species: Optimized for human proteome profiling in metabolic studies.
- Design: Customizable target selection via bioinformatics interface for metabolic pathway analysis.
- Proteins:21- to 384-plex panels targeting adipokines, insulin signaling proteins, lipid metabolism regulators, and inflammatory markers.
- Sample: 1 µL plasma/serum per analysis for minimal sample consumption.
- Readout: NPX-based quantification for normalized protein expression values.
- Platform: PEA-based detection system ensuring high sensitivity and reproducibility.
- Exceptional Sensitivity: Detects low-abundance metabolic regulators (e.g., adipokines, insulin signaling proteins) down to pg/mL concentrations.
- Robust Reproducibility: Delivers consistent results with inter-assay coefficients of variation (CV) below 10% for reliable longitudinal metabolic studies.
- Flexible Panel Options: Offers pre-validated panels targeting metabolic pathways (e.g., glucose homeostasis, lipid metabolism) and fully customizable target selection for specific research needs.
List of Our Metabolic Diseases Proteome Assay Panel
Protein category
The Metabolic Diseases Proteome Assay Panel quantitatively analyzes proteins across six essential functional categories for metabolic research: Nutrient Sensing Receptors, Lipid Metabolism Enzymes, Insulin Signaling Components, Adipokine Signaling Proteins, Inflammatory Mediators, and Mitochondrial Function Regulators (see Table. List of Metabolic Diseases Biomarker Assay).
This panel encompasses fundamental biological processes, including glucose homeostasis, lipid processing, energy balance regulation, metabolic-inflammatory crosstalk, and cellular stress adaptation, featuring strategically selected targets implicated in metabolic pathway dysregulation and compensatory mechanisms. Through systematic literature curation and experimental validation, we incorporate rigorously vetted biomarkers associated with insulin sensitivity, adipocyte function, hepatic metabolism, and pancreatic beta cell communication, providing researchers with a specialized tool for comprehensive proteome investigations in diverse metabolic research areas including obesity studies, diabetes mechanisms, and nutritional intervention research. The panel's optimized design enables focused examination of metabolic disease-related protein interaction networks while preserving the analytical depth required for mechanistic discovery in metabolic biology.
Table. List of Metabolic Diseases Biomarker Assays
Protein Functions
Biological process
primarily involved in key biological processes including nutrient sensing and energy homeostasis regulation, insulin signaling transduction, lipid metabolism and adipokine signaling pathways, inflammatory- metabolic crosstalk mechanisms, and cellular stress adaptation in metabolic tissues, enabling comprehensive investigation of molecular networks governing metabolic function and dysregulation.
Disease area
primarily utilized in research areas involving insulin resistance mechanisms, obesity-related metabolic dysfunction, non-alcoholic fatty liver disease pathogenesis, pancreatic beta cell function studies, and adipose tissue biology, providing researchers with essential tools for investigating molecular pathways underlying metabolic disorders and developing targeted intervention strategies.
The Application of Metabolic Diseases Proteome Assay.
The Metabolic Diseases Proteome Assay enables high-throughput quantification of protein biomarkers across biological pathways, providing researchers with a powerful tool for:
- Discovery of protein networks underlying insulin resistance and lipid metabolism dysregulation in nutritional studies;
- Mechanistic investigation of inflammatory signaling pathways in adipose tissue and hepatic metabolic research;
- Identification of molecular signatures associated with mitochondrial dysfunction and oxidative stress in metabolic models;
- Analysis of adipokine signaling alterations and nutrient-sensing pathway disruptions in obesity research.
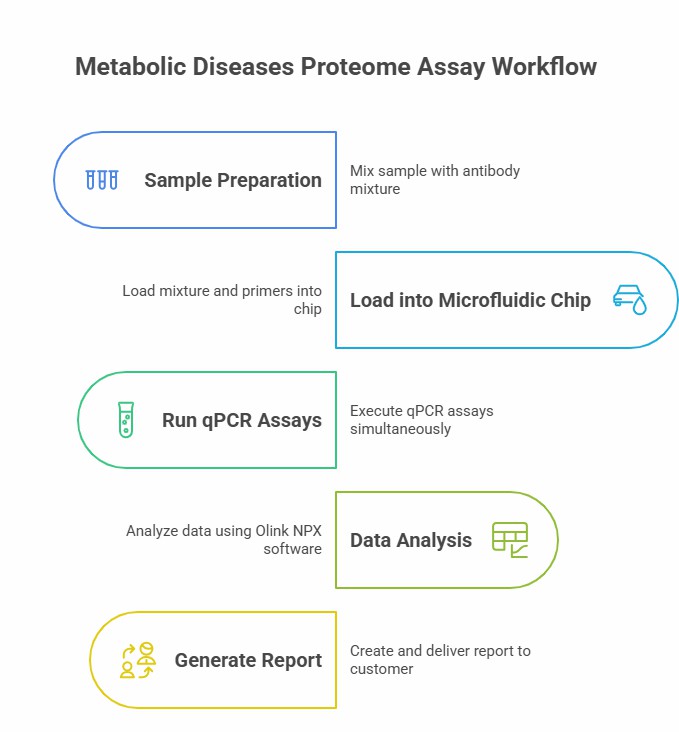
Workflow of Olink Proteomics
Why Creativ Proteomics
Metabolic-Specific Analytical Frameworks
Preconfigured workflows for insulin resistance, lipid metabolism, and adipose inflammation studies with 60+ validated biomarker ratios.
Multi-Omics Integration
Compatible with transcriptomic, metabolomic, and genomic data for systems-level metabolic research (e.g., correlation of protein expression with metabolite shifts).
Pathway-Centric Bioinformatics
Automated tools for enrichment analysis of metabolic pathways (e.g., glycolysis, gluconeogenesis, fatty acid oxidation)
Scalable Throughput
Processes 96–384 samples per run with automation compatibility for large-scale metabolic cohort studies.
Demo Results of Olink Data
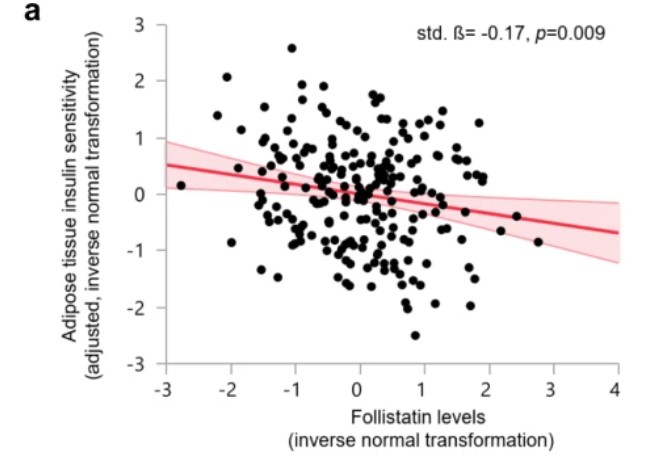 Figure 1: In the Tübingen diabetes family study cohort, the relationship between adipose tissue insulin sensitivity and follistatin levels was analyzed. (Wu, C., et al. 2021)
Figure 1: In the Tübingen diabetes family study cohort, the relationship between adipose tissue insulin sensitivity and follistatin levels was analyzed. (Wu, C., et al. 2021)
Sample Requirements
| Sample Type | Recommended Sample Size | Sample Quality | Pre-treatment and Storage | Sample Transport |
| Plasma/Serum/Body Fluid | The minimum input volume per assay is 40 µL. | For optimal results, maintain concentrations between 0.5 and 1 mg/mL. | For long-term preservation, aliquot samples and maintain at -80°C. | Ship sealed samples on dry ice (-80°C). |
| Tissue | ||||
| Cells | ||||
| Exosomes | ||||
| Other |
Case Study

Short-Term Severe Energy Restriction Promotes Molecular Health and Reverses Aging Signatures in Adults with Prediabetes in the PREVIEW Study
Journal: Aging cell
Year: 2025
- Background
- Methods
- Results
Prediabetes is characterized by impaired fasting blood glucose and/or reduced glucose tolerance, a condition that leads to organ damage, increased mortality, and accelerated aging before diabetes develops. Strict short-term energy restriction while maintaining essential nutrient intake is one of the most effective strategies for weight loss, improved metabolic health, and delaying the progression of type 2 diabetes. Exosomes help achieve these metabolic benefits; However, the effects of weight loss due to energy restriction on the exosomal proteome are still not fully understood.
At the WEHI Biogenomics Laboratory, we performed multiple quantifications of protein biomarkers using proximity extension assay (PEA) using the Olink Targeted Inflammation Kit (96 assays). The kit contains 92 protein assays and 4 internal controls. For this assay, the researchers took plasma samples from the same tubes used for mass spectrometry analysis, and all samples were thawed at the same time. In short, 1 microliter of plasma sample per assay is incubated with a paired set of oligonucleotide-labeled assay antibodies and DNA amplified to generate a specific barcode for each interaction. These barcodes were detected using the Biomark HD instrument and HX controller at 96.96 protein expression IFC. The detection cycle threshold [C(t)] value was then normalized with internal extension control and external plate-to-plate control and processed using Olink tagging software and the OlinkAnalyze R package to convert it to NPX (Standardized Protein Expression) values. These values are expressed in NPX units, displayed on a log-2 scale, while delta-NPX represents the difference between paired longitudinal samples. For the Olink Inflammation Kit, the study used a paired t-test to analyze the differences in protein levels at time points before and after ER. To eliminate the risk of false positives due to multiple comparisons, the Benjamini-Hochberg method was used to adjust the p-value.
To complement the mass spectrometry data, we employ a quantitative detection method for cytokines (Olink Targeted Inflammation Assay Panel) that covers 92 proteins associated with inflammation and senescence to study the effects of energy restriction on inflammatory proteins. Of the 92 proteins analyzed, 75 were detected in more than 75% of the samples. Of these, 29 proteins (32%) showed significant changes after the intervention, with 25 proteins downregulated and 4 proteins upregulated (Figure 1A), indicating an overall reduction in inflammatory markers. The most significant down-regulated proteins based on p-values are shown in Figure 1B.
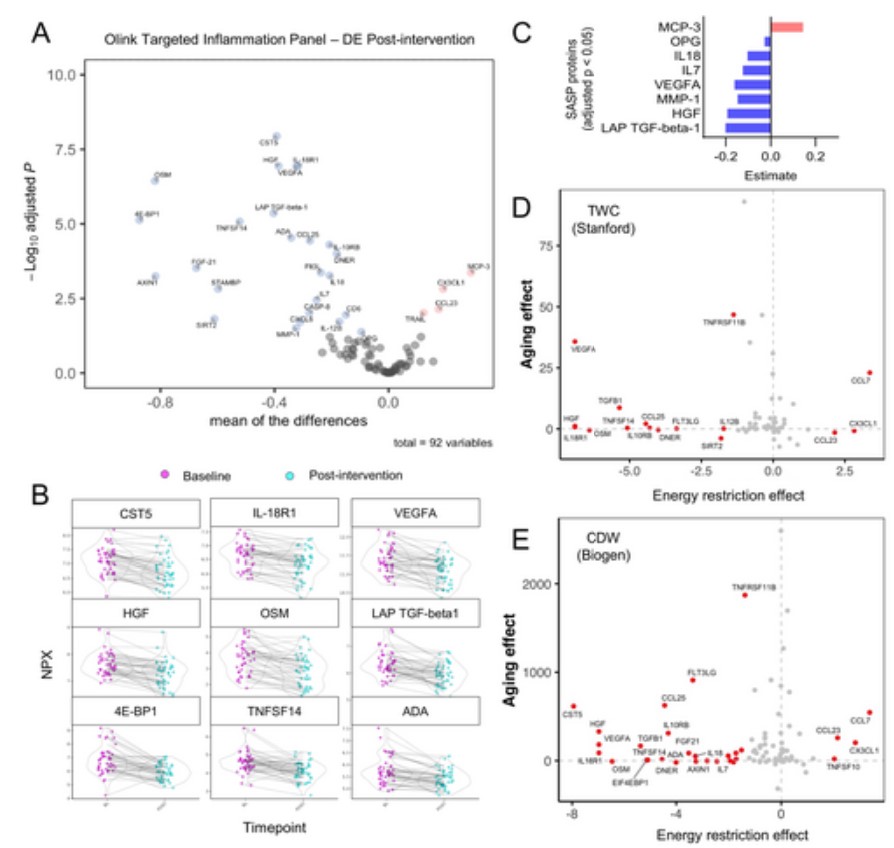 Figure 1. Weight loss due to energy restriction can reduce the content of age-related factors and reverse the aging effects caused by a variety of inflammatory proteins. (Cagigas, M. L et al. 2025)
Figure 1. Weight loss due to energy restriction can reduce the content of age-related factors and reverse the aging effects caused by a variety of inflammatory proteins. (Cagigas, M. L et al. 2025)
FAQs
What research applications does this assay support?
- Investigation of insulin resistance and glucose homeostasis mechanisms
- Lipid metabolism dysregulation studies in obesity models
- Adipose tissue inflammation and adipokine signaling research
- Mitochondrial function analysis in metabolic tissues
What sample types are compatible with the metabolic diseases panel?
The assay supports plasma, serum, and cell culture supernatants, with optimized protocols for each matrix to ensure reproducibility in metabolic studies.
How does the technology achieve high specificity for metabolic biomarkers?
The platform employs PEA technology with dual antibody recognition, enabling precise detection of low-abundance metabolic regulators like adipokines and insulin signaling proteins
What support exists for multi-omics integration?
The platform provides standardized outputs compatible with major omics analysis platforms for systems-level metabolic research.
References
- Wu, C., Borné, Y., Gao, R., et al. (2021). Elevated circulating follistatin associates with an increased risk of type 2 diabetes. Nature communications, 12(1), 6486. https://doi.org/10.1038/s41467-021-26536-w
- Cagigas, M. L., Masedunskas, A., Lin, Y., et al. (2025). Short-Term Severe Energy Restriction Promotes Molecular Health and Reverses Aging Signatures in Adults with Prediabetes in the PREVIEW Study. Aging cell, 24(8), e70123. https://doi.org/10.1111/acel.70123